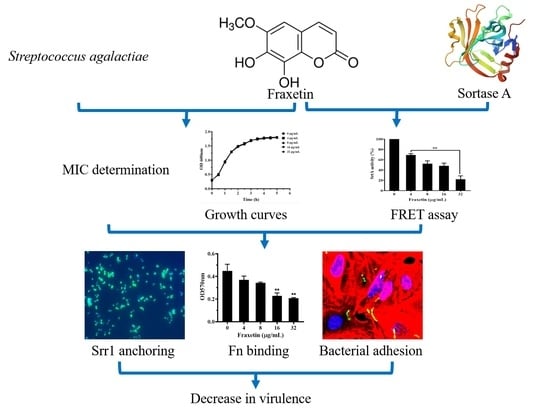
Fraxetin Targeting to Sortase A Decreases the Pathogenicity of Streptococcus agalactiae to Nile Tilapia
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Reagents
2.2. Minimal Inhibitory Concentration (MIC) Determination
2.3. Growth Curves Assay
2.4. Inhibition of SrtA Activity with Fraxetin
2.5. Immunofluorescence Staining of Srr1 on Cell Surface
2.6. Fibronectin Binding Assay
2.7. Bacterial Adhesion Assay
2.8. Challenge Test
2.9. Statistical Analysis
3. Results
3.1. Impact of Fraxetin on Bacterial Growth
3.2. Fraxetin Decreased the Peptidase Activity of SrtA
3.3. Fraxetin Influenced the Anchoring of Srr1 to Cell Surface
3.4. Fraxetin Affected the Binding Ability of S. agalactiae to Fibronectin
3.5. Fraxetin Inhibited the Adhesion of S. agalactiae to A549 Cells
3.6. Fraxetin Protected Tilapia against S. agalactiae Infection
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, G.; Zhu, J.; Chen, K.; Gao, T.; Yao, H.; Liu, Y.; Zhang, W.; Lu, C. Development of Streptococcus agalactiae vaccines for tilapia. Dis. Aquat. Organ. 2016, 122, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, C.A.; Lafrentz, B.R.; Klesius, P.H. Vaccination of sex reversed hybrid tilapia (Oreochromis niloticus × O. aureus) with an inactivated Vibrio vulnificus vaccine. Biologicals 2011, 39, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Mian, G.F.; Godoy, D.T.; Leal, C.A.; Yuhara, T.Y.; Costa, G.M.; Figueiredo, H.C. Aspects of the natural history and virulence of S. agalactiae infection in Nile tilapia. Vet. Microbiol. 2009, 136, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, R.; Li, L.P.; Liang, W.W.; Li, J.; Huang, Y.; Lei, A.Y.; Huang, W.Y.; Gan, X. Screening vaccine candidate strains against Streptococcus agalactiae of tilapia based on PFGE genotype. Vaccine 2012, 30, 6088–6092. [Google Scholar] [CrossRef] [PubMed]
- Depaola, A.; Peller, J.T.; Rodrick, G.E. Effect of oxytetracycline-medicated feed on antibiotic resistance of gram-negative bacteria in catfish ponds. Appl. Environ. Microbiol. 1995, 61, 3513. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Martinez, J.L.; Canton, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Dramsi, S.; Bierne, H. Spatial Organization of Cell Wall-Anchored Proteins at the Surface of Gram-Positive Bacteria. Curr. Top. Microbiol. Immunol. 2017, 404, 177–201. [Google Scholar] [CrossRef] [PubMed]
- Clatworthy, A.E.; Pierson, E.; Hung, D.T. Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007, 3, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Bowmik, S.; Paul, S.K.; Biswas, B.; Banerjee, S.K.; Murty, U.S.; Ravichandiran, V.; Mohan, U. Sortase A: A chemoenzymatic approach for the labeling of cell surfaces. Biotechnol. Bioeng. 2021, 118, 4577–4589. [Google Scholar] [CrossRef]
- Hou, X.C.; Wang, M.N.; Wen, Y.; Ni, T.F.; Guan, X.N.; Lan, L.F.; Zhang, N.X.; Zhang, A.; Yang, C.G. Quinone skeleton as a new class of irreversible inhibitors against Staphylococcus aureus sortase A. Bioorg Med. Chem. Lett. 2018, 28, 1864–1869. [Google Scholar] [CrossRef]
- Alharthi, S.; Alavi, S.E.; Moyle, P.M.; Ziora, Z.M. Sortase A (SrtA) inhibitors as an alternative treatment for superbug infections. Drug Discov. Today 2021, 26, 2164–2172. [Google Scholar] [CrossRef] [PubMed]
- Lalioui, L.; Pellegrini, E.; Dramsi, S.; Baptista, M.; Bourgeois, N.; Doucet-Populaire, F.; Rusniok, C.; Zouine, M.; Glaser, P.; Kunst, F.; et al. The SrtA sortase of Streptococcus agalactiae is required for cell wall anchoring of proteins containing the LPXTG motif, for adhesion to epithelial cells, and for colonization of the mouse intestine. Infect. Immun. 2005, 73, 3342–3350. [Google Scholar] [CrossRef] [PubMed]
- Mistou, M.Y.; Dramsi, S.; Brega, S.; Poyart, C.; Trieu-Cuot, P. Molecular Dissection of the secA2 Locus of Group B Streptococcus Reveals that Glycosylation of the Srr1 LPXTG Protein Is Required for Full Virulence. J. Bacteriol. 2009, 191, 4195–4206. [Google Scholar] [CrossRef]
- Bradshaw, W.J.; Davies, A.H.; Chambers, C.J.; Roberts, A.K.; Shone, C.C.; Acharya, K.R. Molecular features of the sortase enzyme family. FEBS J. 2015, 282, 2097–2114. [Google Scholar] [CrossRef]
- Mosihuzzaman, M. Herbal Medicine in Healthcare-An Overview. Nat. Prod. Commun. 2012, 7, 807–812. [Google Scholar] [CrossRef]
- Reverter, M.; Bontemps, N.; Lecchini, D.; Banaigs, B.; Sasal, P. Use of plant extracts in fish aquaculture as an alternative to chemotherapy: Current status and future perspectives. Aquaculture 2014, 433, 50–61. [Google Scholar] [CrossRef]
- Zhang, W.N.; Zhao, J.P.; Ma, Y.F.; Li, J.; Chen, X.H. The effective components of herbal medicines used for prevention and control of fish diseases. Fish. Shellfish. Immun. 2022, 126, 73–83. [Google Scholar] [CrossRef]
- Witaicenis, A.; Seito, L.N.; Chagas, A.D.; de Almeida, L.D.; Luchini, A.C.; Rodrigues-Orsi, P.; Cestari, S.H.; Di Stasi, L.C. Antioxidant and intestinal anti-inflammatory effects of plant-derived coumarin derivatives. Phytomedicine 2014, 21, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.T.; Zou, D.; Xie, K.P.; Xie, M.J. Antibacterial mechanism of fraxetin against Staphylococcus aureus. Mol. Med. Rep. 2014, 10, 2341–2345. [Google Scholar] [CrossRef]
- Dong, J.; Zhang, L.S.; Xu, N.; Zhou, S.; Song, Y.; Yang, Q.H.; Liu, Y.T.; Yang, Y.B.; Ai, X.H. Rutin reduces the pathogenicity of Streptococcus agalactiae to tilapia by inhibiting the activity of sortase A. Aquaculture 2021, 530, 735743. [Google Scholar] [CrossRef]
- CLSI. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd ed.; Approved Standard (M45-ED3); CLSI: Wayne, PA, USA, 2015. [Google Scholar]
- Wang, J.F.; Li, H.E.; Pan, J.; Dong, J.; Zhou, X.; Niu, X.D.; Deng, X.M. Oligopeptide Targeting Sortase A as Potential Anti-infective Therapy for Staphylococcus aureus. Front. Microbiol. 2018, 9, 245. [Google Scholar] [CrossRef]
- Six, A.; Bellais, S.; Bouaboud, A.; Fouet, A.; Gabriel, C.; Tazi, A.; Dramsi, S.; Trieu-Cuot, P.; Poyart, C. Srr2, a multifaceted adhesin expressed by ST-17 hypervirulent Group B Streptococcus involved in binding to both fibrinogen and plasminogen. Mol. Microbiol. 2015, 97, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- Sudheesh, P.S.; Crane, S.; Cain, K.D.; Strom, M.S. Sortase inhibitor phenyl vinyl sulfone inhibits Renibacterium salmoninarum adherence and invasion of host cells. Dis. Aquat. Org. 2007, 78, 115–127. [Google Scholar] [CrossRef]
- Cho, E.; Hwang, J.Y.; Park, J.S.; Oh, D.; Oh, D.C.; Park, H.G.; Shin, J.; Oh, K.B. Inhibition of Streptococcus mutans adhesion and biofilm formation with small-molecule inhibitors of sortase A from Juniperus chinensis. J. Oral. Microbiol. 2022, 14, 2088937. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.M.R.M.; Mitra, S.; Bin Emran, T.; Dhama, K.; Ripon, M.K.H.; Gajdacs, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Chen, J.M.; Sun, R.X.; Pan, C.G.; Sun, Y.; Mai, B.X.; Li, Q.X. Antibiotics and Food Safety in Aquaculture. J. Agric. Food Chem. 2020, 68, 11908–11919. [Google Scholar] [CrossRef] [PubMed]
- Au, M.B.; Haserick, J.R.; Chen, Y.S.; Gibson, F.C.; Deng, L.L. Structural Identification of Lipid-a: A Glycosyl Lipid Involved in Oligo- and Polysaccharides Metabolism in Streptococcus agalactiae (Group B Streptococcus). Curr. Microbiol. 2023, 80, 16. [Google Scholar] [CrossRef] [PubMed]
- Raabe, V.N.; Shane, A.L. Group B Streptococcus (Streptococcus agalactiae). Microbiol. Spectr. 2019, 7, GPP3-0007-2018. [Google Scholar] [CrossRef]
- Savini, V.; Santarelli, A.; Argentieri, A.V.; D‘Amario, C.; Fazii, P.; D‘Antonio, D. Group B Streptococcus agalactiae interspecies exchange. Vet. Microbiol. 2013, 165, 487–488. [Google Scholar] [CrossRef]
- Zhang, Z.; Lan, J.F.; Li, Y.H.; Hu, M.Q.; Yu, A.G.; Zhang, J.; Wei, S. The pathogenic and antimicrobial characteristics of an emerging Streptococcus agalactiae serotype IX in Tilapia. Microb. Pathog. 2018, 122, 39–45. [Google Scholar] [CrossRef]
- Geng, Y.; Wang, K.Y.; Huang, X.L.; Chen, D.F.; Li, C.W.; Ren, S.Y.; Liao, Y.T.; Zhou, Z.Y.; Liu, Q.F.; Du, Z.J.; et al. Streptococcus agalactiae, an Emerging Pathogen for Cultured Ya-Fish, Schizothorax prenanti, in China. Transbound. Emerg. Dis. 2012, 59, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Alazab, A.; Sadat, A.; Younis, G. Prevalence, antimicrobial susceptibility, and genotyping of Streptococcus agalactiae in Tilapia fish (Oreochromis niloticus) in Egypt. J. Adv. Vet. Anim. Res. 2022, 9, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xie, K.; Zou, D.; Zhou, L.; Xie, M. A preliminary study: Inhibitory mechanism of fraxetin on Escherichia coli. Chin. J. Microecol. 2014, 26, 1123–1126. [Google Scholar]
- Zhu, L.; Chen, K.; Xi, B.; Xie, J. In vitro antibacterial effect of fraxetin on pathogenic Aeromonas hydrophila. J. Fish. Sci. China 2019, 26, 984–992. [Google Scholar]
- Liu, L.; Wang, R.; Chen, L.; Yang, Q.; Weng, X.; Sun, J. Comparative study on antibacterial action of coumarin monomer and ash bark from different origins. Chin. J. Inf. Tradit. Chin. Med. 2009, 16, 39–42. [Google Scholar]
- Wang, J.; Song, M.; Pan, J.; Shen, X.; Liu, W.; Zhang, X.; Li, H.; Deng, X. Quercetin impairs Streptococcus pneumoniae biofilm formation by inhibiting sortase A activity. J. Cell Mol. Med. 2018, 22, 6228–6237. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shi, Y.; Jing, S.; Dong, H.; Wang, D.; Wang, T. Astilbin Inhibits the Activity of Sortase A from Streptococcus mutans. Molecules 2019, 24, 465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Deng, Y.; Wang, X.; Shi, L.; Zhan, B.; Hou, N.; Liu, S.; Bao, M.; Chi, G.; Fang, T. Alnustone inhibits Streptococcus pneumoniae virulence by targeting pneumolysin and sortase A. Fitoterapia 2022, 162, 105261. [Google Scholar] [CrossRef]
- Hu, P.; Huang, P.; Chen, M.W. Curcumin reduces Streptococcus mutans biofilm formation by inhibiting sortase A activity. Arch. Oral. Biol. 2013, 58, 1343–1348. [Google Scholar] [CrossRef]
- Xu, Y.F.; Lin, H.Y.; Wang, H.; Pang, J.; Zhou, Y. Fraxetin attenuates ferroptosis in myocardial infarction via AKT/Nrf2/HO-1 signaling. Am. J. Transl. Res. 2021, 13, 10315–10327. [Google Scholar]
- Shi, Y.J.; Sun, Z.Y.; Liu, Y.; Shu, J.Y.; Zhang, Y.; Lv, Q.H.; Wang, J.F.; Deng, X.M.; Liu, H.T.; Qiu, J.Z. Inhibition of the Type III Secretion System of Salmonella enterica Serovar Typhimurium via Treatment with Fraxetin. Microbiol. Spectr. 2022, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L. Study on the Anti-Infective Mechanism of Fraxetin on Aeromonas hydrophila; Shanghai Ocean University: Shanghai, China, 2021. [Google Scholar]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, J.; Zhang, Y.; Yang, Q.; Liu, Y.; Zhou, S.; Ai, X. Fraxetin Targeting to Sortase A Decreases the Pathogenicity of Streptococcus agalactiae to Nile Tilapia. Animals 2024, 14, 1337. https://doi.org/10.3390/ani14091337
Dong J, Zhang Y, Yang Q, Liu Y, Zhou S, Ai X. Fraxetin Targeting to Sortase A Decreases the Pathogenicity of Streptococcus agalactiae to Nile Tilapia. Animals. 2024; 14(9):1337. https://doi.org/10.3390/ani14091337
Chicago/Turabian StyleDong, Jing, Yuze Zhang, Qiuhong Yang, Yongtao Liu, Shun Zhou, and Xiaohui Ai. 2024. "Fraxetin Targeting to Sortase A Decreases the Pathogenicity of Streptococcus agalactiae to Nile Tilapia" Animals 14, no. 9: 1337. https://doi.org/10.3390/ani14091337