DReAmocracy: A Method to Capitalise on Prior Drug Discovery Efforts to Highlight Candidate Drugs for Repurposing
Abstract
:1. Introduction
2. Results
2.1. Reference Tables for Each Disease, Type of Analysis and Drug Feature
2.2. Common Signatures in CTS vs. CDRS for Neurodegenerative Diseases
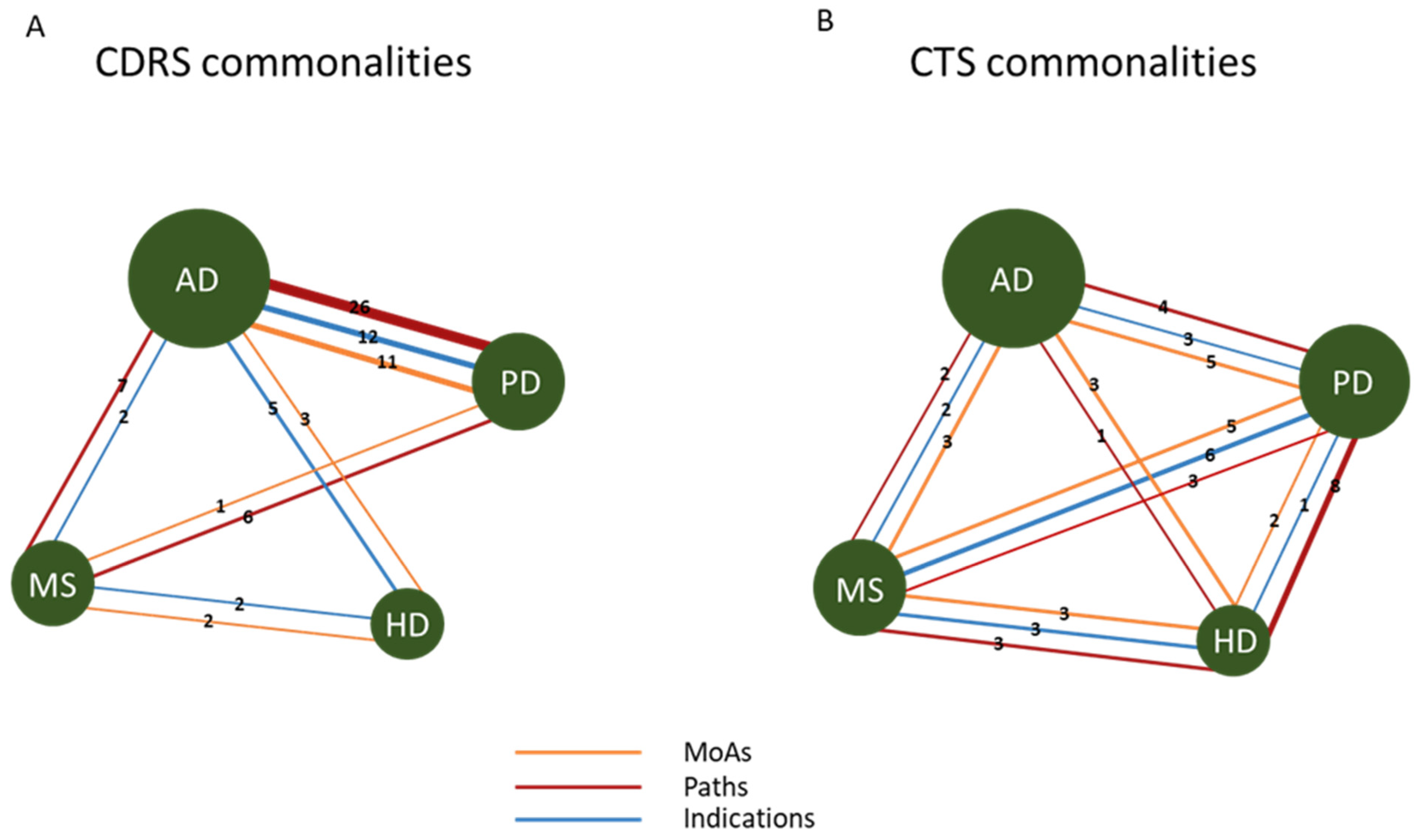
2.3. Commonalities and Differences in Signatures across Neurodegenerative Diseases
2.4. Generation of the Super-Reference Table of Drugs
2.5. Scoring New Candidate Drugs for Repurposing against a Selected Disease
3. Discussion
4. Materials and Methods
4.1. Data Selection
4.2. Construction of the Reference Score Matrix (DC-Matrix)
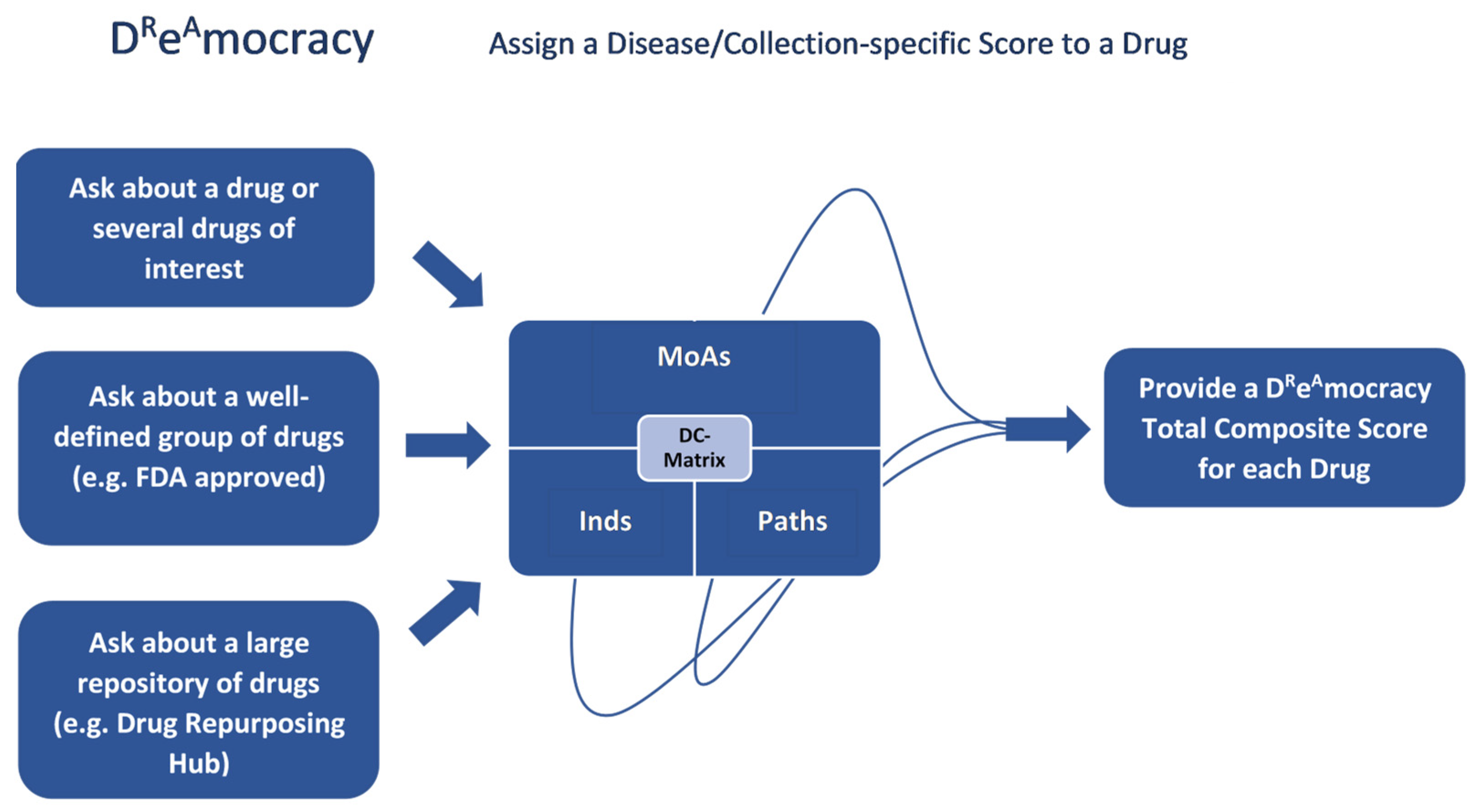
4.3. Assign a Disease/Collection-Specific Score to a Drug
4.4. Drug Repurposing Hub through the lens of DReAmocracy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s Disease |
| ALL | Acute lymphoblastic leukaemia |
| ALS | Amyotrophic Lateral Sclerosis |
| AMPK | AMP-activated Protein Kinase |
| CDRS | Computational drug repurposing studies |
| CMap | Connectivity Map |
| CTCL | T-cell lymphoma |
| CTS | Clinical trial studies |
| DC Score | Drug/Collection-specific Score |
| EIB | Exercise-induced bronchoconstriction |
| FDA | Food and Drug Administration |
| HD | Huntington’s Disease |
| HMGCR | 3-hydroxy-3-methylglutaryl-CoA reductase |
| Ind | Initial indication |
| MoA | Mechanism of action |
| MS | Multiple Sclerosis |
| Path | Pathway |
| PD | Parkinson’s Disease |
| PPAR | Peroxisome Proliferator-activated Receptors |
| VMAT2 | Vesicular monoamine transporter 2 |
References
- Himmelstein, D.S.; Lizee, A.; Hessler, C.; Brueggeman, L.; Chen, S.L.; Hadley, D.; Green, A.; Khankhanian, P.; Baranzini, S.E.; University of California; et al. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. Elife 2017, 6, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Kropiwnicki, E.; Lachmann, A.; Clarke, D.J.B.; Xie, Z.; Jagodnik, K.M.; Ma’ayan, A. DrugShot: Querying biomedical search terms to retrieve prioritized lists of small molecules. BMC Bioinform. 2022, 23, 76. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Che, C.; Jin, B.; Zhang, N.; Su, C.; Wang, F. Knowledge-driven drug repurposing using a comprehensive drug knowledge graph. Health Inform. J. 2020, 26, 2737–2750. [Google Scholar] [CrossRef] [PubMed]
- Bai, Q.; Liu, S.; Tian, Y.; Xu, T.; Banegas-Luna, A.J.; Pérez-Sánchez, H.; Huang, J.; Liu, H.; Yao, X. Application advances of deep learning methods for de novo drug design and molecular dynamics simulation. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2022, 12, e1581. [Google Scholar] [CrossRef]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2018, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Iorio, F.; Bosotti, R.; Scacheri, E.; Belcastro, V.; Mithbaokar, P.; Ferriero, R.; Murino, L.; Tagliaferri, R.; Brunetti-Pierri, N.; Isacchi, A.; et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl. Acad. Sci. USA 2010, 107, 14621–14626. [Google Scholar] [CrossRef] [PubMed]
- Gloeckner, C.; Garner, A.L.; Mersha, F.; Oksov, Y.; Tricoche, N.; Eubanks, L.M.; Lustigman, S.; Kaufmann, G.F.; Janda, K.D. Repositioning of an existing drug for the neglected tropical disease Onchocerciasis. Proc. Natl. Acad. Sci. USA 2010, 107, 3424–3429. [Google Scholar] [CrossRef] [PubMed]
- Keiser, M.J.; Setola, V.; Irwin, J.J.; Laggner, C.; Abbas, A.I.; Hufeisen, S.J.; Jensen, N.H.; Kuijer, M.B.; Matos, R.C.; Tran, T.B.; et al. Predicting new molecular targets for known drugs. Nature 2009, 462, 175–181. [Google Scholar] [CrossRef]
- Dudley, J.T.; Deshpande, T.; Butte, A.J. Exploiting drug-disease relationships for computational drug repositioning. Brief. Bioinform. 2011, 12, 303–311. [Google Scholar] [CrossRef]
- Lal, J.C.; Mao, C.; Zhou, Y.; Gore-Panter, S.R.; Rennison, J.H.; Lovano, B.S.; Castel, L.; Shin, J.; Gillinov, A.M.; Smith, J.D.; et al. Transcriptomics-based network medicine approach identifies metformin as a repurposable drug for atrial fibrillation. Cell Rep. Med. 2022, 3, 100749. [Google Scholar] [CrossRef] [PubMed]
- Panda, P.K.; Arul, M.N.; Patel, P.; Verma, S.K.; Luo, W.; Rubahn, H.-G.; Mishra, Y.K.; Suar, M.; Ahuja, R. Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci. Adv. 2020, 6, eabb8097. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, S.; Prema, K.V.; Balaji, S. Machine learning models for drug–target interactions: Current knowledge and future directions. Drug Discov. Today 2020, 25, 748–756. [Google Scholar] [CrossRef]
- Wang, H.; Guo, F.; Du, M.; Wang, G.; Cao, C. A novel method for drug-target interaction prediction based on graph transformers model. BMC Bioinform. 2022, 23, 459. [Google Scholar] [CrossRef] [PubMed]
- Yasar, S.; Xia, J.; Yao, W.; Furberg, C.D.; Xue, Q.-L.; Mercado, C.I.; Fitzpatrick, A.L.; Fried, L.P.; Kawas, C.H.; Sink, K.M.; et al. Antihypertensive drugs decrease risk of Alzheimer disease Ginkgo Evaluation of Memory Study. Neurology 2013, 81, 896–903. [Google Scholar] [CrossRef]
- Adesuyan, M.; Jani, Y.; Alsugeir, D.; Cheung, E.; Chu, C.; Howard, R.; Wong, I.; Brauer, R. Antihypertensive Agents and Incident Alzheimer’s Disease: A Systematic Review and Meta-Analysis of Observational Studies. J. Prev. Alzheimer’s Dis. 2022, 9, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Davis-Plourde, K.L.; Sedaghat, S.; Tully, P.J.; Wang, W.; Phillips, C.; Pase, M.P.; Himali, J.J.; Windham, B.G.; Griswold, M.; et al. Antihypertensive medications and risk for incident dementia and Alzheimer’s disease: A meta-analysis of individual participant data from prospective cohort studies. Lancet Neurol. 2020, 19, 61–70. [Google Scholar] [CrossRef]
- Toal, C.B.; Meredith, P.A.; Elliott, H.L. Long-acting dihydropyridine calcium-channel blockers and sympathetic nervous system activity in hypertension: A literature review comparing amlodipine and nifedipine GITS. Blood Press 2012, 21 (Suppl. S1), 3–10. [Google Scholar] [CrossRef] [PubMed]
- Goto, S. Striatal Gαolf/cAMP signal-dependent mechanism to generate levodopa-induced dyskinesia in Parkinson’s disease. Front. Cell. Neurosci. 2017, 11, 364. [Google Scholar] [CrossRef]
- Fjodorova, M.; Noakes, Z.; De La Fuente, D.C.; Errington, A.C.; Li, M. Dysfunction of cAMP–Protein Kinase A–Calcium Signaling Axis in Striatal Medium Spiny Neurons: A Role in Schizophrenia and Huntington’s Disease Neuropathology. Biol. Psychiatry Glob. Open Sci. 2022, 3, 418–429. [Google Scholar] [CrossRef]
- Hartung, H.P.; Gonsette, R.; Konig, N.; Kwiecinski, H.; Guseo, A.; Morrissey, S.P.; Krapf, H.; Zwingers, T. Mitoxantrone in progressive multiple sclerosis: A placebo- controlled, double-blind, randomised, multicentre trial. Lancet 2018, 360, 2018–2025. [Google Scholar] [CrossRef] [PubMed]
- Dunk, A.; Scott, S.; Johnson, P.; Melia, W.; Lok, A.; Murray-Lyon, I.; Williams, R.; Thomas, H. Mitozantrone as single agent therapy in hepatocellular carcinoma. A phase II study. J. Hepatol. 1985, 1, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elrahman, K.S.; Hamilton, A.; Hutchinson, S.R.; Liu, F.; Russell, R.C.; Ferguson, S.S.G. mGluR5 antagonism increases autophagy and prevents disease progression in the zQ175 mouse model of Huntington’s disease. Sci. Signal. 2017, 10, eaan6387. [Google Scholar] [CrossRef] [PubMed]
- Morsali, D.; Bechtold, D.; Lee, W.; Chauhdry, S.; Palchaudhuri, U.; Hassoon, P.; Snell, D.M.; Malpass, K.; Piers, T.; Pocock, J.; et al. Safinamide and flecainide protect axons and reduce microglial activation in models of multiple sclerosis. Brain 2013, 136, 1067–1082. [Google Scholar] [CrossRef] [PubMed]
- Andrews, M.; Tousi, B.; Sabbagh, M.N. 5HT6 Antagonists in the Treatment of Alzheimer’s Dementia: Current Progress. Neurol. Ther. 2018, 7, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Padala, K.P.; Padala, P.R.; McNeilly, D.P.; Geske, J.A.; Sullivan, D.H.; Potter, J.F. The effect of HMG-CoA reductase inhibitors on cognition in patients with alzheimer’s dementia: A prospective withdrawal and rechallenge pilot study. Am. J. Geriatr. Pharmacother. 2012, 10, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Marucci, G.; Buccioni, M.; Ben, D.D.; Lambertucci, C.; Volpini, R.; Amenta, F. Efficacy of acetylcholinesterase inhibitors in Alzheimer’s disease. Neuropharmacology 2021, 190, 108352. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Rowe, M.; Ren, M.; Hong, J.-S.; Chen, P.-S.; Chuang, D.-M. Histone deacetylase inhibitors exhibit anti-inflammatory and neuroprotective effects in a rat permanent ischemic model of stroke: Multiple mechanisms of action. J. Pharmacol. Exp. Ther. 2007, 321, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.-S.; Li, G.; Tzeng, N.-S.; Chen, P.-S.; Chuang, D.-M.; Hsu, Y.-D.; Yang, S.; Hong, J.-S. Valproate pretreatment protects dopaminergic neurons from LPS-induced neurotoxicity in rat primary midbrain cultures: Role of microglia. Mol. Brain Res. 2005, 134, 162–169. [Google Scholar] [CrossRef]
- Sáiz-Vázquez, O.; Gracia-García, P.; Ubillos-Landa, S.; Puente-Martínez, A.; Casado-Yusta, S.; Olaya, B.; Santabárbara, J. Depression as a Risk Factor for Alzheimer’ s Disease: A Systematic Review of Longitudinal Meta-Analyses. J. Clin. Med. 2021, 10, 1809. [Google Scholar] [CrossRef]
- Walczak-Nowicka, Ł.J.; Herbet, M. Acetylcholinesterase inhibitors in the treatment of neurodegenerative diseases and the role of acetylcholinesterase in their pathogenesis. Int. J. Mol. Sci. 2021, 22, 9290. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, X.; Li, Y.; Tang, T.S.; Bezprozvanny, I. Tetrabenazine is neuroprotective in Huntington’s disease mice. Mol. Neurodegener. 2010, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Foo, E.; Russell, M.; Lily, O.; Ford, H. Mitoxantrone in relapsing-remitting and rapidly progressive multiple sclerosis: Ten-year clinical outcomes post-treatment with mitoxantrone. Mult. Scler. Relat. Disord. 2020, 44, 102330. [Google Scholar] [CrossRef]
- Barbagallo, M. Type 2 diabetes mellitus and Alzheimer’s disease. World J. Diabetes 2014, 5, 889. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, Y.-J.; Jang, Y.; Kwon, Y.H. AMPK activation inhibits apoptosis and tau hyperphosphorylation mediated by palmitate in SH-SY5Y cells. Brain Res. 2011, 1418, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.P.; Feng, L.; Yap, K.B.; Lee, T.S.; Tan, C.H.; Winblad, B. Long-term metformin usage and cognitive function among older adults with diabetes. J. Alzheimer’s Dis. 2014, 41, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Gannon, M.; Che, P.; Chen, Y.; Jiao, K.; Roberson, E.D.; Wang, Q. Noradrenergic dysfunction in Alzheimer’s disease. Front. Neurosci. 2015, 9, 220. [Google Scholar] [CrossRef] [PubMed]
- Delaville, C.; De Deurwaerdère, P.; Benazzouz, A. Noradrenaline and Parkinson’s disease. Front. Syst. Neurosci. 2011, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Miranda, A.S.; Cardozo, P.L.; Silva, F.R.; de Souza, J.M.; Olmo, I.G.; Cruz, J.S.; Gomez, M.V.; Ribeiro, F.M.; Vieira, L.B. Alterations of Calcium Channels in a Mouse Model of Huntington’ s Disease and Neuroprotection by Blockage of CaV1 Channels. ASN Neuro 2019, 11, 1–11. [Google Scholar] [CrossRef]
- Masato, A.; Plotegher, N.; Boassa, D.; Bubacco, L. Impaired dopamine metabolism in Parkinson’s disease pathogenesis. Mol. Neurodegener. 2019, 14, 35. [Google Scholar] [CrossRef]
- Nobili, A.; Latagliata, E.C.; Viscomi, M.T.; Cavallucci, V.; Cutuli, D.; Giacovazzo, G.; Krashia, P.; Rizzo, F.R.; Marino, R.; Federici, M.; et al. Dopamine neuronal loss contributes to memory and reward dysfunction in a model of Alzheimer’s disease. Nat. Commun. 2017, 8, 14727. [Google Scholar] [CrossRef] [PubMed]
- Savva, K.; Zachariou, M.; Bourdakou, M.M.; Dietis, N.; Spyrou, G.M. Network-based stage-specific drug repurposing for Alzheimer ’ s disease. Comput. Struct. Biotechnol. J. 2022, 20, 1427–1438. [Google Scholar] [CrossRef] [PubMed]
- Nicolai, L.; Nettesheim, P.; de Witte, L.D.; Snijders, G. Antidepressants as a potential candidate to reduce microglia activation in neurodegenerative diseases. A systematic review and meta-analysis of preclinical studies. J. Affect. Disord. Rep. 2023, 11, 100465. [Google Scholar] [CrossRef]
- Zhang, G.; Stackman, R.W. The role of serotonin 5-HT2A receptors in memory and cognition. Front. Pharmacol. 2015, 6, 225. [Google Scholar] [CrossRef] [PubMed]
- Steventon, J.J.; Rosser, A.E.; Hart, E.; Murphy, K. Hypertension, Antihypertensive Use and the Delayed-Onset of Huntington’s Disease. Mov. Disord. 2020, 35, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, S.; Vatsa, N.; Kumar, V.; Singh, B.K.; Jamal, I.; Sharma, A.; Jana, N.R. Topoisomerase 1 inhibitor topotecan delays the disease progression in a mouse model of Huntington’s disease. Hum. Mol. Genet. 2017, 26, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, R.; Nizzari, M.; Zanardi, I.; Pusch, M.; Gavazzo, P. Voltage-Gated Sodium Channel Dysfunctions in Neurological Disorders. Life 2023, 13, 1191. [Google Scholar] [CrossRef] [PubMed]
- Al-Rafiah, A.; Magadmi, R.; Al-Kaabi, A.; Alsomali, N. Parkinson’s Disease-Related Biomarkers That May Appear in Amphetamine Abusers. Biomed. Res. Int. 2021, 2021, 3081891. [Google Scholar] [CrossRef]
- Chen, L.; Yu, P.; Zhang, L.; Zou, Y.; Zhang, Y.; Jiang, L.; Gao, R.; Xiao, H.; Qian, Y.; Wang, J. Methamphetamine exposure induces neuropathic protein β-Amyloid expression. Toxicol. Vitr. 2019, 54, 304–309. [Google Scholar] [CrossRef]
- Xu, H.; Chen, X.; Wang, J.; Yang, T.; Liu, N.; Cheng, J.; Gao, R.; Liu, J.; Xiao, H. Involvement of insulin signalling pathway in methamphetamine-induced hyperphosphorylation of Tau. Toxicology 2018, 408, 88–94. [Google Scholar] [CrossRef]
- Wang, R.; Reddy, H. Role of Glutamate and NMDA in Alzheimer’s disease. J. Alzheimer’s Dis. 2017, 57, 1041–1048. [Google Scholar] [CrossRef]
- Olianas, M.C.; Dedoni, S.; Onali, P. The atypical antidepressant mianserin exhibits agonist activity at κ-opioid receptors. Br. J. Pharmacol. 2012, 167, 1329–1341. [Google Scholar] [CrossRef] [PubMed]
- Mullard, A. Symptomatic AD treatment fails in first phase III. Nat. Rev. Drug Discov. 2016, 15, 738. [Google Scholar] [CrossRef] [PubMed]
- Teodorescu, A.; Dima, L.; Ifteni, P.; Rogozea, L.M. Clozapine for Treatment-Refractory Behavioral Disturbance in Dementia. Am. J. Ther. 2018, 25, E320–E325. [Google Scholar] [CrossRef] [PubMed]
- Dinesh, M.; David, A.; Quraishi, S.N. Depot pipotiazine palmitate and undecylenate for schizophrenia. Cochrane Database Syst. Rev. 2009. [Google Scholar] [CrossRef]
- Kropiwnicki, E.; Evangelista, J.E.; Stein, D.J.; Clarke, D.J.B.; Lachmann, A.; Kuleshov, M.V.; Jeon, M.; Jagodnik, K.M.; Ma’ayan, A. Drugmonizome and Drugmonizome-ML: Integration and abstraction of small molecule attributes for drug enrichment analysis and machine learning. Database 2021, 2021, baab017. [Google Scholar] [CrossRef]







| Drug Name | AD Total Composite Score | Drug Name | PD Total Composite Score | Drug Name | HD Total Composite Score | Drug Name | MS Total Composite Score |
|---|---|---|---|---|---|---|---|
| agomelatine | 0.85 | apomorphine | 0.73 | chlorpromazine | 0.68 | zonisamide | 0.55 |
| mirtazapine | 0.85 | pramipexole | 0.73 | fluphenazine | 0.68 | disopyramide | 0.52 |
| vortioxetine | 0.83 | lisuride | 0.72 | perphenazine | 0.68 | priralfimide | 0.52 |
| aripiprazole | 0.83 | terguride | 0.70 | trifluoperazine | 0.68 | dalfampridine | 0.51 |
| mianserin | 0.80 | bromocriptine | 0.70 | risperidone | 0.63 | chloroprocaine | 0.49 |
| sarpogrelate | 0.77 | ropinirole | 0.68 | pimozide | 0.63 | valproic-acid | 0.48 |
| cyamemazine | 0.76 | rotigotine | 0.68 | chlorprothixene | 0.58 | cinchocaine | 0.46 |
| clozapine | 0.74 | piribedil | 0.68 | clozapine | 0.58 | ibutilide | 0.45 |
| loxapine | 0.73 | fenoldopam | 0.67 | flupentixol | 0.58 | haloperidol–decanoate | 0.45 |
| ketanserin | 0.73 | mirtazapine | 0.66 | iloperidone | 0.58 | troglitazone | 0.44 |
| methysergide | 0.72 | a-412997 | 0.64 | levomepromazine | 0.58 | spironolactone | 0.42 |
| quetiapine | 0.72 | abt-724 | 0.64 | olanzapine | 0.58 | tesaglitazar | 0.42 |
| ziprasidone | 0.72 | etilevodopa | 0.64 | pipotiazine | 0.58 | rosiglitazone | 0.42 |
| blonserin | 0.72 | ro-10-5824 | 0.64 | pipotiazine–palmitate | 0.58 | pioglitazone | 0.42 |
| paliperidone | 0.72 | metixene | 0.64 | promazine | 0.58 | flibanserin | 0.41 |
| gr-113808 | 0.72 | agomelatine | 0.64 | spiperone | 0.58 | phenacemide | 0.40 |
| gr125487 | 0.72 | mesulergine | 0.63 | thioproperazine | 0.58 | eslicarbazepine–acetate | 0.40 |
| idalopirdine | 0.72 | talipexole | 0.62 | thiothixene | 0.58 | oxcarbazepine | 0.40 |
| r-1485 | 0.72 | dr-4485 | 0.62 | zuclopenthixol | 0.58 | procaine | 0.40 |
| rs-23597-190 | 0.72 | sb-269970 | 0.62 | amisulpride | 0.57 | amiloride | 0.39 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savva, K.; Zachariou, M.; Bourdakou, M.M.; Dietis, N.; Spyrou, G.M. DReAmocracy: A Method to Capitalise on Prior Drug Discovery Efforts to Highlight Candidate Drugs for Repurposing. Int. J. Mol. Sci. 2024, 25, 5319. https://doi.org/10.3390/ijms25105319
Savva K, Zachariou M, Bourdakou MM, Dietis N, Spyrou GM. DReAmocracy: A Method to Capitalise on Prior Drug Discovery Efforts to Highlight Candidate Drugs for Repurposing. International Journal of Molecular Sciences. 2024; 25(10):5319. https://doi.org/10.3390/ijms25105319
Chicago/Turabian StyleSavva, Kyriaki, Margarita Zachariou, Marilena M. Bourdakou, Nikolas Dietis, and George M. Spyrou. 2024. "DReAmocracy: A Method to Capitalise on Prior Drug Discovery Efforts to Highlight Candidate Drugs for Repurposing" International Journal of Molecular Sciences 25, no. 10: 5319. https://doi.org/10.3390/ijms25105319







