Antibody–Drug Conjugate Made of Zoledronic Acid and the Anti-CD30 Brentuximab–Vedotin Exert Anti-Lymphoma and Immunostimulating Effects
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis and Chemical Characterization of Bre–Ved–ZA ADC
2.2. Flow Cytometry and Confocal Microscopy
2.3. Evaluation of HL-Cell-Line Proliferation and Apoptosis
2.4. Vδ2 T-Cell Proliferation
2.5. Cytotoxicity Assays
2.6. Statistical Analysis
3. Results
3.1. Production, Chemical Characterization, and Reactivity of Bre–Ved–ZA ADC
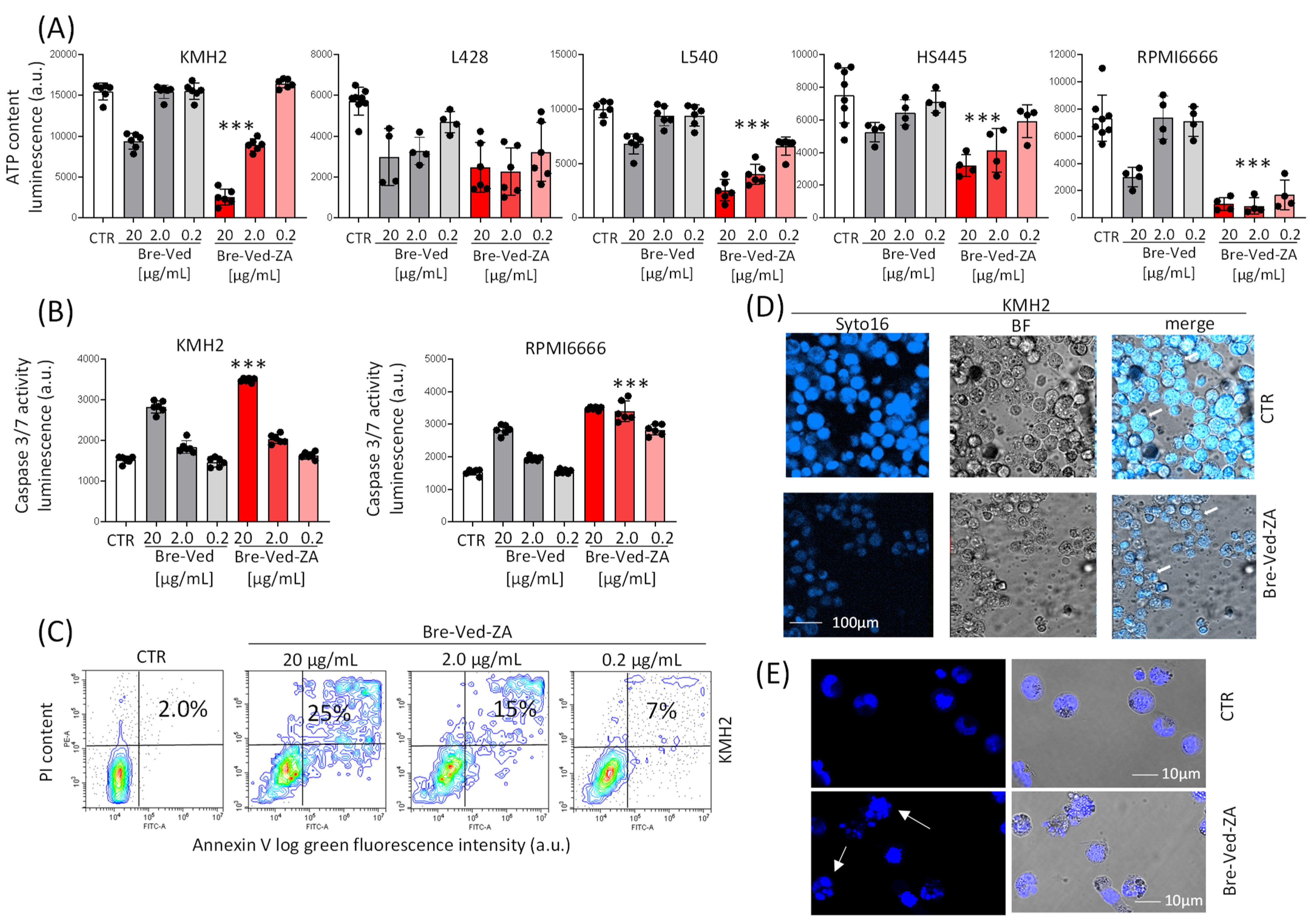
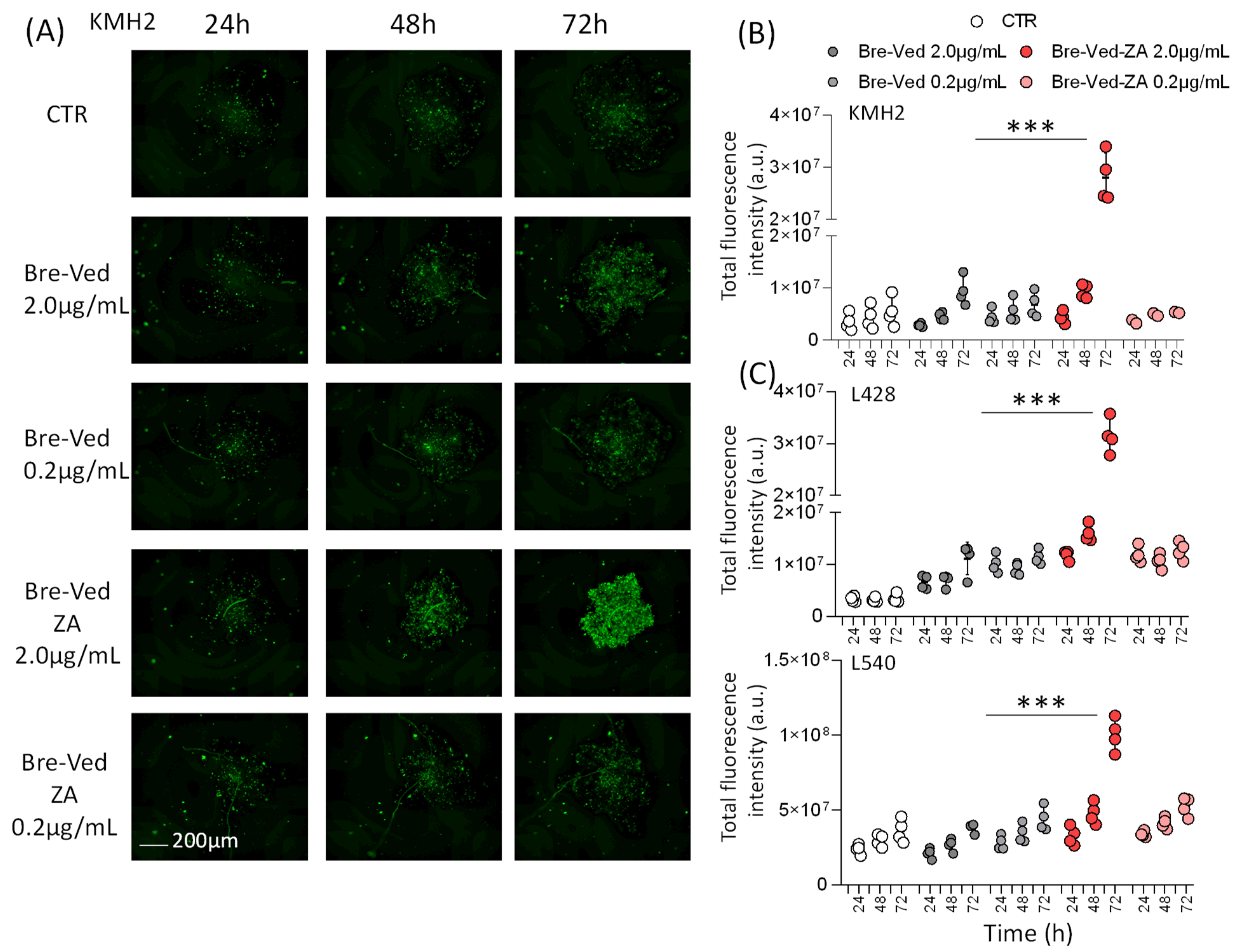
3.2. Bre–Ved–ZA ADC Localizes in the Lysosomal Compartment and Interferes with HL-Cell Growth
3.3. Bre–Ved–ZA ADC Can Stimulate Effector Vδ2 T-Cells with Antitumor Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fridman, W.H.; Pages, F.; Sautes-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Church, S.E.; Galon, J. Tumor Microenvironment and immunotherapy: The whole picture is better than a glimpse. Immunity 2015, 43, 631–633. [Google Scholar] [CrossRef] [PubMed]
- Bonneville, M.; O’Brien, R.L.; Born, W.K. γδ T cell effector functions: A blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 2010, 10, 467–478. [Google Scholar] [CrossRef]
- Wang, C.Q.; Lim, P.Y.; Tan, A.H. Gamma/delta T cells as cellular vehicles for anti-tumor immunity. Front. Immunol. 2024, 14, 1282758. [Google Scholar] [CrossRef] [PubMed]
- Bhat, J.; Kabelitz, D. γδ T cells and epigenetic drugs: A useful merger in cancer immunotherapy? Oncoimmunology 2015, 4, e1006088. [Google Scholar] [CrossRef] [PubMed]
- Kabelitz, D.; Wesch, D.; He, W. Perspectives of γδ T cells in tumor immunology. Cancer Res. 2007, 67, 5–8. [Google Scholar] [CrossRef]
- Vavassori, S.; Kumar, A.; Wan, G.S.; Ramanjaneyulu, G.S.; Cavallari, M.; El Daker, S.; Beddoe, T.; Theodossis, A.; Williams, N.K.; Gostick, E.; et al. Butyrophilin 3A1 binds phosphorylated antigens and stimulates human γδ T cells. Nat. Immunol. 2013, 14, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Cano, C.E.; Pasero, C.; De Gassart, A.; Kerneur, C.; Gabriac, M.; Fullana, M.; Granarolo, E.; Hoet, R.; Scotet, E.; Rafia, C.; et al. BTN2A1, an immune checkpoint targeting Vγ9Vδ2 T cell cytotoxicity against malignant cells. Cell Rep. 2021, 36, 109359. [Google Scholar] [CrossRef]
- Morita, C.T.; Jin, C.; Sarikonda, G.; Wang, H. Nonpeptide antigens, presentation mechanisms, and immunological memory of human Vγ2Vδ2 T cells: Discriminating friend from foe through the recognition of prenyl pyrophosphate antigens. Immunol. Rev. 2007, 215, 59–76. [Google Scholar] [CrossRef] [PubMed]
- Di Mascolo, D.; Varesano, S.; Benelli, R.; Mollica, H.; Salis, A.; Zocchi, M.R.; Decuzzi, P.; Poggi, A. Nanoformulated Zoledronic Acid Boosts the Vδ2 T Cell Immunotherapeutic Potential in Colorectal Cancer. Cancers 2020, 12, 104. [Google Scholar] [CrossRef]
- Santini, D.; Vespasiani Gentilucci, U.; Vincenzi, B.; Picardi, A.; Vasaturo, F.; La Cesa, A.; Onori, N.; Scarpa, S.; Tonini, G. The antineoplastic role of bisphosphonates: From basic research to clinical evidence. Ann. Oncol. 2003, 14, 1468–1476. [Google Scholar] [CrossRef] [PubMed]
- Santolaria, T.; Robard, M.; Léger, A.; Catros, V.; Bonneville, M.; Scotet, E. Repeated systemic administration of aminobisphosphonates and human Vγ9Vδ2 T cells efficiently control tumor development in vivo. J. Immunol. 2013, 191, 1993–2000. [Google Scholar] [CrossRef]
- Arnett, H.A.; Viney, J.L. Immune modulation by butyrophilins. Nat. Rev. Immunol. 2014, 14, 559–569. [Google Scholar] [CrossRef]
- Tosetti, F.; Venè, R.; Camodeca, C.; Nuti, E.; Rossello, A.; D’Arrigo, C.; Galante, D.; Ferrari, N.; Poggi, A.; Zocchi, M.R. Specific ADAM10 inhibitors localize in exosome-like vesicles released by Hodgkin lymphoma and stromal cells and prevent sheddase activity carried to bystander cells. OncoImmunology 2018, 7, e1421889. [Google Scholar] [CrossRef] [PubMed]
- Pece, R.; Tavella, S.; Costa, D.; Varesano, S.; Camodeca, C.; Cuffaro, D.; Nuti, E.; Rossello, A.; Alfano, M.; D’Arrigo, C.; et al. Inhibitors of ADAM10 reduce Hodgkin lymphoma cell growth in 3D microenvironments and enhance brentuximab-vedotin effect. Haematologica 2022, 107, 909–920. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Scott, L.J. BrentuximabVedotin: A Review in CD30-Positive Hodgkin Lymphoma. Drugs 2017, 77, 435–445. [Google Scholar] [CrossRef]
- Lynch, R.C.; Cassaday, R.D.; Smith, S.D.; Fromm, J.R.; Cowan, A.J.; Warren, E.H.; Shadman, M.S.; Shustov, A.; Till, B.G.; Ujjani, C.S.; et al. Dose-dense brentuximab vedotin plus ifosfamide, carboplatin, and etoposide for second-line treatment of relapsed or refractory classical Hodgkin lymphoma: A single centre, phase 1/2 study. Lancet Haematol. 2021, 8, e562–e571. [Google Scholar] [CrossRef]
- LaCasce, A.S. Treating Hodgkin lymphoma in the new millennium: Relapsed and refractory disease. Hematol. Oncol. 2019, 37 (Suppl. S1), 87–91. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Bertozzi, C.R. Site-specific antibody-drug conjugates: The nexus of bioorthogonal chemistry, protein engineering, and drug development. Bioconjugate Chem. 2015, 26, 176–192. [Google Scholar] [CrossRef]
- Baah, S.; Laws, M.; Rahman, K.M. Antibody–Drug Conjugates-A Tutorial Review. Molecules 2021, 26, 2943. [Google Scholar] [CrossRef]
- Dean, A.Q.; Luo, S.; Twomey, J.D.; Zhang, B. Targeting cancer with antibody-drug conjugates: Promises and challenges. mAbs 2021, 13, e1951427. [Google Scholar] [CrossRef] [PubMed]
- Benelli, R.; Costa, D.; Salvini, L.; Tardito, S.; Tosetti, F.; Villa, F.; Zocchi, M.R.; Poggi, A. Targeting of colorectal cancer organoids with zoledronic acid conjugated to the anti-EGFR antibody cetuximab. J. ImmunoTherapy Cancer 2022, 10, e005660. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.S.; Kao, P.M.; McCue, A.W.; Chappelle, H.L. Use of maleimide-thiol coupling chemistry for efficient syntheses of oligonucleotide-enzyme conjugate hybridization probes. Bioconjugate Chem. 1990, 1, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Panowski, S.; Bhakta, S.; Raab, H.; Polakis, P.; Junutula, J.R. Site-specific antibody drug conjugates for cancer therapy. mAbs 2014, 6, 34–45. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shabarova, Z.A.; Ivanovskaya, M.G.; Isaguliants, M.G. DNA-like duplexes with repetitions: Efficient template-guided polycondensation of decadeoxyribonucleotide imidazolide. FEBS Lett. 1983, 154, 288–292. [Google Scholar] [CrossRef]
- Itumoh, E.J.; Data, S.; Leitao, E.M. Opening up the Toolbox: Synthesis and Mechanisms of Phosphoramidates. Molecules 2020, 25, 3684. [Google Scholar] [CrossRef]
- Nakashima, M.; Uchimaru, K. CD30 Expression and Its Functions during the Disease Progression of Adult T-Cell Leukemia/Lymphoma. Int. J. Mol. Sci. 2023, 24, 8731. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Faber, M.L.; Oldham, R.A.A.; Thakur, A.; Rademacher, M.J.; Kubicka, E.; Dlugi, T.A.; Gifford, S.A.; McKillop, W.M.; Schloemer, N.J.; Lum, L.G.; et al. Novel anti-CD30/CD3 bispecific antibodies activate human T cells and mediate potent anti-tumor activity. Front. Immunol. 2023, 14, 1225610. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Brice, P.; de Kerviler, E.; Friedberg, J.W. Classical Hodgkin lymphoma. Lancet 2021, 398, 1518–1527. [Google Scholar] [CrossRef] [PubMed]
- Kahn, J.; Dabaja, B.; Wu, S.; Kelly, K.; Berkahn, L.; Pavlovsky, A.; Sureda, A.; LaCasce, A. Classic Hodgkin lymphoma. Hematol. Oncol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Schimmoeller, C.J.; Bastian, C.; Fleming, J.; Morales, J. A Review of Hodgkin Lymphoma in the Era of Checkpoint Inhibitors. Cureus. 2023, 15, e41660. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tamma, R.; Ingravallo, G.; Gaudio, F.; d’Amati, A.; Masciopinto, P.; Bellitti, E.; Lorusso, L.; Annese, T.; Benagiano, V.; Musto, P.; et al. The Tumor Microenvironment in Classic Hodgkin’s Lymphoma in Responder and No-Responder Patients to First Line ABVD Therapy. Cancers 2023, 15, 2803. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stewart, B.J.; Fergie, M.; Young, M.D.; Jones, C.; Sachdeva, A.; Blain, A.; Bacon, C.M.; Rand, V.; Ferdinand, J.R.; James, K.R.; et al. Spatial and molecular profiling of the mononuclear phagocyte network in classic Hodgkin lymphoma. Blood 2023, 141, 2343–2358. [Google Scholar] [CrossRef] [PubMed]
- Ferrarini, I.; Bernardelli, A.; Lovato, E.; Schena, A.; Krampera, M.; Visco, C. An updated portrait of monocyte-macrophages in classical Hodgkin lymphoma. Front. Oncol. 2023, 13, 1149616. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Beck, A.; Goetsch, L.; Dumontet, C.; Corvaïa, N. Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug Discov. 2017, 16, 315–337. [Google Scholar] [CrossRef] [PubMed]
- Tsuchikama, K.; Anami, Y.; Ha, S.Y.Y.; Yamazaki, C.M. Exploring the next generation of antibody-drug conjugates. Nat. Rev. Clin. Oncol. 2024, 21, 203–223. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Li, P.; Yang, T.; Zhu, J.; Sun, L.; Zhang, Z.; Wang, L.; Tian, X.; Chen, J.; Hu, C.; et al. The promise and challenges of combination therapies with antibody-drug conjugates in solid tumors. J. Hematol. Oncol. 2024, 17, 1. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sapra, P.; Stein, R.; Pickett, J.; Qu, Z.; Govindan, S.V.; Cardillo, T.M.; Hansen, H.J.; Horak, I.D.; Griffiths, G.L.; Goldenberg, D.M. Anti-CD74 antibody-doxorubicin conjugate, IMMU-110, in a human multiple myeloma xenograft and in monkeys. Clin. Cancer Res. 2005, 11, 5257–5264. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.G.; Vrabel, M.R.; Mantooth, S.M.; Hopkins, J.J.; Wagner, E.S.; Gabaldon, T.A.; Zaharoff, D.A. Localized Interleukin-12 for Cancer Immunotherapy. Front. Immunol. 2020, 11, 575597. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dumontet, C.; Reichert, J.M.; Senter, P.D.; Lambert, J.M.; Beck, A. Antibody-drug conjugates come of age in oncology. Nat Rev Drug Discov. 2023, 8, 641–661. [Google Scholar] [CrossRef] [PubMed]
- Cianferotti, C.; Faltoni, V.; Cini, E.; Ermini, E.; Migliorini, F.; Petricci, E.; Taddei, M.; Salvini, L.; Battistuzzi, G.; Milazzo, F.M.; et al. Antibody drug conjugates with hydroxamic acid cargos for histone deacetylase (HDAC) inhibition. Chem. Commun. 2021, 57, 867–870. [Google Scholar] [CrossRef] [PubMed]
- Khattak, Z.E.; Hashmi, H.; Khan, S.I.; Aamir, S.; Arif, U.; Khan, A.I.; Darwin, A.; Singh, A.D.; Khouri, J.; Anwer, F. Dawn of a new era of antibody-drug conjugates and bispecific T-cell engagers for treatment of multiple myeloma: A systematic review of literature. Ann. Hematol. 2021, 100, 2155–2172. [Google Scholar] [CrossRef] [PubMed]
- List, T.; Casi, G.; Neri, D. A chemically defined trifunctional antibody-cytokine-drug conjugate with potent antitumor activity. Mol. Cancer Ther. 2014, 13, 2641–2652. [Google Scholar] [CrossRef] [PubMed]
- Olatunji, F.P.; Herman, J.W.; Kesic, B.N.; Olabode, D.; Berkman, C.E. A click-ready pH-triggered phosphoramidate-based linker for controlled-release of monomethyl auristatin E. Tetrahedron Lett. 2020, 61, 152398. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, M.; Nakaseko, C.; Ohwada, C.; Takeuchi, M.; Ozawa, S.; Shimizu, N.; Cho, R.; Nishimura, M.; Saito, Y. Zoledronate has an antitumor effect and induces actin rearrangement in dexamethasone-resistant myeloma cells. Eur. J. Haematol. 2007, 79, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, M.; Wang, C.; Zhu, L.; Hu, J.; Chen, S.; Wu, X.; Li, B.; Li, Y. The feature of distribution and clonality of TCR γ/δ subfamilies T cells in patients with B-cell non-Hodgkin lymphoma. J. Immunol. Res. 2014, 2014, 241246. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zocchi, M.R.; Catellani, S.; Canevali, P.; Tavella, S.; Garuti, A.; Villaggio, B.; Zunino, A.; Gobbi, M.; Fraternali-Orcioni, G.; Kunkl, A.; et al. High ERp5/ADAM10 expression in lymph node microenvironment and impaired NKG2D ligands recognition in Hodgkin lymphomas. Blood 2012, 119, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Biswas, P.; Rovere, P.; De Filippi, C.; Heltai, S.; Smith, C.; Dagna, L.; Poli, G.; Manfredi, A.A.; Ferrarini, M. Engagement of CD30 shapes the secretion of cytokines by human γ δ T cells. Eur. J. Immunol. 2000, 30, 2172–2180. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Myneni, S.; Batt, A.; Guerrero, J.; Brumm, J.; Chung, S. Immunogenicity risk assessment for biotherapeutics through in vitro detection of CD134 and CD137 on T helper cells. mAbs 2021, 13, 1898831. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Romano, A.; Parrinello, N.L.; Chiarenza, A.; Motta, G.; Tibullo, D.; Giallongo, C.; La Cava, P.; Camiolo, G.; Puglisi, F.; Palumbo, G.A.; et al. Immune off-target effects of Brentuximab Vedotin in relapsed/refractory Hodgkin Lymphoma. Br. J. Haematol. 2019, 185, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Hansen, H.P.; Paes Leme, A.F.; Hallek, M. Role of ADAM10 as a CD30 Sheddase in Classical Hodgkin Lymphoma. Front. Immunol. 2020, 11, 398. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, Z.; Guo, W.; Bai, O. Mechanism of action and therapeutic targeting of CD30 molecule in lymphomas. Front. Oncol. 2023, 13, 1301437. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Oryan, A.; Sahvieh, S. Effects of bisphosphonates on osteoporosis: Focus on zoledronate. Life Sci. 2021, 264, 118681. [Google Scholar] [CrossRef] [PubMed]






| Average Mass (Daltons) | Sum Intensity | Charge State Distribution | Delta Mass | Assignment | |
|---|---|---|---|---|---|
| LC | 23,724 | 4.47 × 107 | 10–19 | 0 | LC |
| 23,898 | 3.90 × 106 | 10–19 | 174 | +ZA | |
| 25,040 | 2.84 × 107 | 10–17 | 1316 | +Vedotin | |
| 25,214 | 5.06 × 106 | 10–18 | 1490 | +Vedotin + ZA | |
| HC | 50,321 | 2.97 × 107 | 17–37 | 0 | HC glycosylated |
| 50,483 | 1.86 × 107 | 20–34 | 162 | +Hexose | |
| 50,495 | 4.48 × 106 | 20–32 | 174 | +ZA | |
| 51,638 | 3.63 × 107 | 20–37 | 1317 | +Vedotin | |
| 51,800 | 2.45 × 107 | 20–37 | 1479 | +Vedotin + Hexose | |
| 51,812 | 7.80 × 106 | 21–43 | 1491 | +Vedotin + ZA | |
| 51,975 | 6.93 × 106 | 21–43 | 1654 | +Vedotin + Hexose + ZA | |
| 52,192 | 5.29 × 106 | 22–41 | 1871 | +2 Vedotin − 1 MMAE | |
| 52,354 | 5.54 × 106 | 21–36 | 2033 | +2 Vedotin − 1 MMAE + Hexose | |
| 52,955 | 2.30 × 107 | 20–35 | 2634 | +2 Vedotin | |
| 53,116 | 1.30 × 107 | 20–34 | 2795 | +2 Vedotin + Hexose | |
| 53,131 | 3.80 × 106 | 20–37 | 2810 | +2 Vedotin + ZA | |
| 53,509 | 7.11 × 106 | 24–35 | 3188 | +3 Vedotin − 1 MMAE | |
| 53,671 | 5.51 × 106 | 23–38 | 3350 | +3 Vedotin − 1 MMAE + Hexose | |
| 54,271 | 1.61 × 107 | 20–39 | 3950 | +3 Vedotin | |
| 54,432 | 8.81 × 106 | 20–34 | 4111 | +3 Vedotin + Hexose | |
| 54,444 | 5.70 × 106 | 20–33 | 4123 | +3 Vedotin + ZA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morelli, F.; Matis, S.; Benelli, R.; Salvini, L.; Zocchi, M.R.; Poggi, A. Antibody–Drug Conjugate Made of Zoledronic Acid and the Anti-CD30 Brentuximab–Vedotin Exert Anti-Lymphoma and Immunostimulating Effects. Cells 2024, 13, 862. https://doi.org/10.3390/cells13100862
Morelli F, Matis S, Benelli R, Salvini L, Zocchi MR, Poggi A. Antibody–Drug Conjugate Made of Zoledronic Acid and the Anti-CD30 Brentuximab–Vedotin Exert Anti-Lymphoma and Immunostimulating Effects. Cells. 2024; 13(10):862. https://doi.org/10.3390/cells13100862
Chicago/Turabian StyleMorelli, Feliciana, Serena Matis, Roberto Benelli, Laura Salvini, Maria Raffaella Zocchi, and Alessandro Poggi. 2024. "Antibody–Drug Conjugate Made of Zoledronic Acid and the Anti-CD30 Brentuximab–Vedotin Exert Anti-Lymphoma and Immunostimulating Effects" Cells 13, no. 10: 862. https://doi.org/10.3390/cells13100862






