Transcriptome Analysis Reveals Potential Regulators of DMI Fungicide Resistance in the Citrus Postharvest Pathogen Penicillium digitatum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Growth Conditions
2.2. Transcriptome Analysis
2.3. Quantitative Real-Time PCR
2.4. Disruption of P. digitatum Genes and Mutant Complementation
2.5. Mycelial Growth Tests
2.6. Virulence Assays
2.7. Determination of Intracellular Ergosterol Content
3. Results
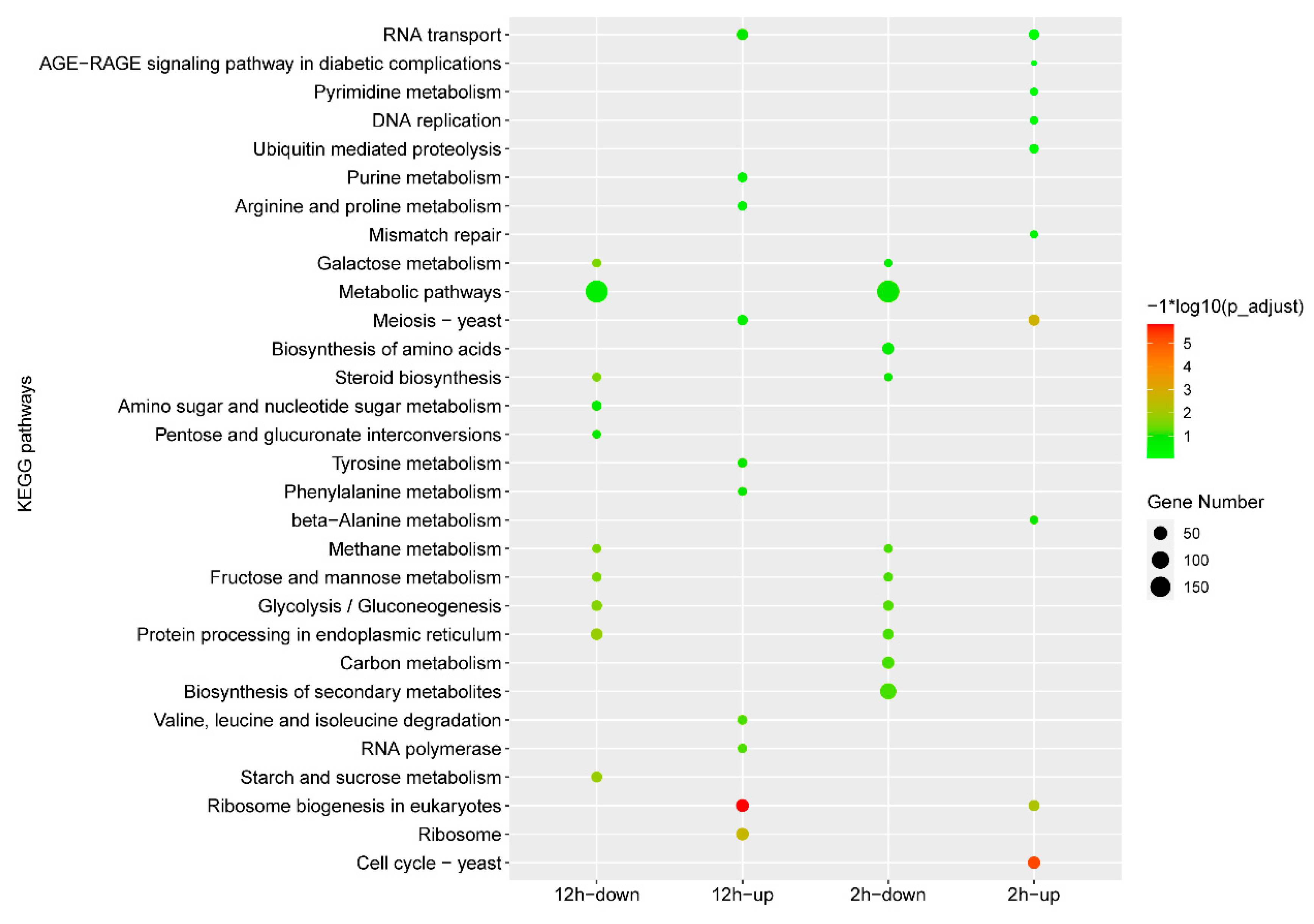
3.1. Transcriptomic Analysis of P. digitatum in Response to IMZ
3.2. Expression of the Ergosterol Biosynthesis Pathway
3.3. Expression of Potential Regulators
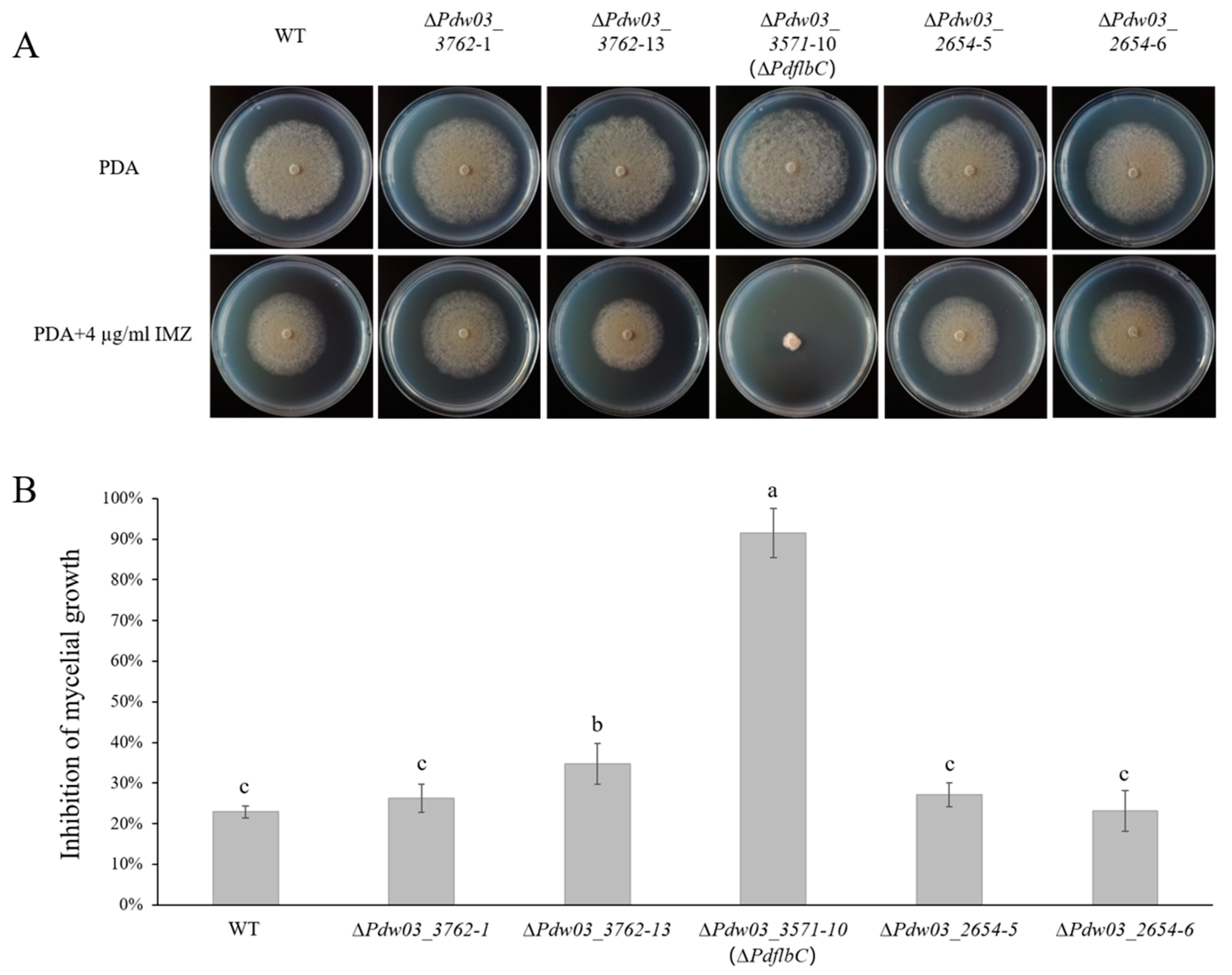
3.4. Disruption of Three Transcription Factors in P. digitatum
3.5. Roles of PdflbC in Regulating Ergosterol Biosynthesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maria, C.S.; Giuseppe, A.; Naouel, A.; Francesco, G.; Giovanni, C.D.R. Advance in Citrus Postharvest Management: Diseases, Cold Storage and Quality Evaluation. In Citrus Pathology; Harsimran, G., Harsh, G., Eds.; IntechOpen: Rijeka, Croatia, 2017; Chapter 7. [Google Scholar]
- Kanetis, L.; Förster, H.; Adaskaveg, J.E. Comparative efficacy of the new postharvest fungicides azoxystrobin, fludioxonil, and pyrimethanil for managing citrus green mold. Plant Dis. 2007, 91, 1502–1511. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Torres, P. Molecular Mechanisms Underlying Fungicide Resistance in Citrus Postharvest Green Mold. J. Fungi 2021, 7, 783. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, Y.; Schnabel, G.; Peng, C.A.; Lagishetty, S.; Smith, K.; Luo, C.; Yuan, H. Inherent Resistance to 14α-Demethylation Inhibitor Fungicides in Colletotrichum truncatum Is Likely Linked to CYP51A and/or CYP51B Gene Variants. Phytopathology 2018, 108, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Carter, H.E.; Fraaije, B.A.; West, J.S.; Kelly, S.L.; Mehl, A.; Shaw, M.W.; Cools, H.J. Alterations in the predicted regulatory and coding regions of the sterol 14α-demethylase gene (CYP51) confer decreased azole sensitivity in the oilseed rape pathogen Pyrenopeziza brassicae. Mol. Plant Pathol. 2014, 15, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, T.M.; Berg, G.; Semaškienė, R.; Mehl, A.; Sierotzki, H.; Stammler, G.; Justesen, A.F.; Jørgensen, L.N. Impact of DMI and SDHI fungicides on disease control and CYP51 mutations in populations of Zymoseptoria tritici from Northern Europe. Eur. J. Plant Pathol. 2015, 143, 861–871. [Google Scholar] [CrossRef]
- Mair, W.J.; Deng, W.; Mullins, J.G.; West, S.; Wang, P.; Besharat, N.; Ellwood, S.R.; Oliver, R.P.; Lopez-Ruiz, F.J. Demethylase inhibitor fungicide resistance in Pyrenophora teres f. sp. teres associated with target site modification and inducible overexpression of Cyp51. Front. Microbiol. 2016, 7, 1279. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Du, J.; Chi, M.; Sun, X.; Liang, W.; Huang, J.; Li, B. The Y137H mutation in the cytochrome P450 FgCYP51B protein confers reduced sensitivity to tebuconazole in Fusarium graminearum. Pest Manag. Sci. 2018, 74, 1472–1477. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, G.; Jones, A.L. The 14α-demethylasse (CYP51A1) gene is overexpressed in Venturia inaequalis strains resistant to myclobutanil. Phytopathology 2001, 91, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Proffer, T.J.; Jacobs, J.L.; Sundin, G.W. Overexpression of the 14α-demethylase target gene (CYP51) mediates fungicide resistance in Blumeriella jaapii. Appl. Environ. Microbiol. 2006, 72, 2581–2585. [Google Scholar] [CrossRef]
- Luo, C.-X.; Cox, K.D.; Amiri, A.; Schnabel, G. Occurrence and detection of the DMI resistance-associated genetic element ‘Mona’ in Monilinia fructicola. Plant Dis. 2008, 92, 1099–1103. [Google Scholar] [CrossRef]
- Diaz-Trujillo, C.; Chong, P.; Stergiopoulos, I.; Cordovez, V.; Guzman, M.; De Wit, P.J.; Meijer, H.J.; Scalliet, G.; Sierotzki, H.; Lilia Peralta, E. A new mechanism for reduced sensitivity to demethylation-inhibitor fungicides in the fungal banana black Sigatoka pathogen Pseudocercospora fijiensis. Mol. Plant Pathol. 2018, 19, 1491–1503. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Verweij, P.E.; Melchers, W.J.G.; Moye-Rowley, W.S. Differential Functions of Individual Transcription Factor Binding Sites in the Tandem Repeats Found in Clinically Relevant cyp51A Promoters in Aspergillus fumigatus. mBio 2022, 13, e00702–e00722. [Google Scholar] [CrossRef] [PubMed]
- Mernke, D.; Dahm, S.; Walker, A.-S.; Lalève, A.; Fillinger, S.; Leroch, M.; Hahn, M. Two promoter rearrangements in a drug efflux transporter gene are responsible for the appearance and spread of multidrug resistance phenotype MDR2 in Botrytis cinerea isolates in French and German vineyards. Phytopathology 2011, 101, 1176–1183. [Google Scholar] [CrossRef] [PubMed]
- Omrane, S.; Sghyer, H.; Audéon, C.; Lanen, C.; Duplaix, C.; Walker, A.S.; Fillinger, S. Fungicide efflux and the MgMFS 1 transporter contribute to the multidrug resistance phenotype in Zymoseptoria tritici field isolates. Environ. Microbiol. 2015, 17, 2805–2823. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, H.; Hasegawa, K.; Nakaune, R.; Lee, Y.J.; Makizumi, Y.; Akutsu, K.; Hibi, T. Tandem Repeat of a Transcriptional Enhancer Upstream of the Sterol 14α-Demethylase Gene (CYP51) in Penicillium digitatum. Appl. Environ. Microbiol. 2000, 66, 3421–3426. [Google Scholar] [CrossRef] [PubMed]
- Ghosoph, J.M.; Schmidt, L.S.; Margosan, D.A.; Smilanick, J.L. Imazalil resistance linked to a unique insertion sequence in the PdCYP51 promoter region of Penicillium digitatum. Postharvest Biol. Technol. 2007, 44, 9–18. [Google Scholar] [CrossRef]
- Sun, X.; Wang, J.; Feng, D.; Ma, Z.; Li, H. PdCYP51B, a new putative sterol 14alpha-demethylase gene of Penicillium digitatum involved in resistance to imazalil and other fungicides inhibiting ergosterol synthesis. Appl. Microbiol. Biotechnol. 2011, 91, 1107–1119. [Google Scholar] [CrossRef] [PubMed]
- de Ramón-Carbonell, M.; Sánchez-Torres, P. Significance of 195 bp-enhancer of PdCYP51B in the acquisition of Penicillium digitatum DMI resistance and increase of fungal virulence. Pestic. Biochem. Physiol. 2020, 165, 104522. [Google Scholar] [CrossRef] [PubMed]
- Espenshade, P.J.; Hughes, A.L. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 2007, 41, 401–427. [Google Scholar] [CrossRef]
- Bien, C.M.; Espenshade, P.J. Sterol regulatory element binding proteins in fungi: Hypoxic transcription factors linked to pathogenesis. Eukaryot. Cell 2010, 9, 352–359. [Google Scholar] [CrossRef]
- Goldstein, J.L.; DeBose-Boyd, R.A.; Brown, M.S. Protein sensors for membrane sterols. Cell 2006, 124, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Todd, B.L.; Espenshade, P.J. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 2005, 120, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.; Barker, B.M.; Carey, C.C.; Merriman, B.; Werner, E.R.; Lechner, B.E.; Dhingra, S.; Cheng, C.; Xu, W.; Blosser, S.J. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Path. 2014, 10, e1004487. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Chung, H.; Lee, G.-W.; Koh, S.-K.; Chae, S.-K.; Lee, Y.-H. Genome-wide analysis of hypoxia-responsive genes in the rice blast fungus, Magnaporthe oryzae. PLoS ONE 2015, 10, e0134939. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-N.; Wu, F.-Y.; Tian, R.-Y.; Shi, Y.-X.; Xu, Z.-Q.; Liu, J.-Y.; Huang, J.; Xue, F.-F.; Liu, B.-Y.; Liu, G.-Q. The bHLH-zip transcription factor SREBP regulates triterpenoid and lipid metabolisms in the medicinal fungus Ganoderma lingzhi. Commun. Biol. 2023, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Ruan, R.; Wang, M.; Liu, X.; Sun, X.; Chung, K.-R.; Li, H. Functional analysis of two sterol regulatory element binding proteins in Penicillium digitatum. PLoS ONE 2017, 12, e0176485. [Google Scholar] [CrossRef]
- Guida, A.; Lindstädt, C.; Maguire, S.L.; Ding, C.; Higgins, D.G.; Corton, N.J.; Berriman, M.; Butler, G. Using RNA-seq to determine the transcriptional landscape and the hypoxic response of the pathogenic yeast Candida parapsilosis. BMC Genom. 2011, 12, 628. [Google Scholar] [CrossRef] [PubMed]
- Synnott, J.M.; Guida, A.; Mulhern-Haughey, S.; Higgins, D.G.; Butler, G. Regulation of the hypoxic response in Candida albicans. Eukaryot. Cell 2010, 9, 1734–1746. [Google Scholar] [CrossRef] [PubMed]
- Zavrel, M.; Hoot, S.J.; White, T.C. Comparison of sterol import under aerobic and anaerobic conditions in three fungal species: Candida albicans, Candida glabrata and Saccharomyces cerevisiae. Eukaryot. Cell 2013, 12, 725–738. [Google Scholar] [CrossRef]
- Rogers, P.D. Sterol homeostasis in yeast. Nat. Chem. Biol. 2022, 18, 1170–1171. [Google Scholar] [CrossRef]
- Tan, L.; Chen, L.; Yang, H.; Jin, B.; Kim, G.; Im, Y.J. Structural basis for activation of fungal sterol receptor Upc2 and azole resistance. Nat. Chem. Biol. 2022, 18, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Jian, Y.; Chen, Y.; Kistler, H.C.; He, P.; Ma, Z.; Yin, Y. A phosphorylated transcription factor regulates sterol biosynthesis in Fusarium graminearum. Nat. Commun. 2019, 10, 1228. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ruan, R.; Li, H. The completed genome sequence of the pathogenic ascomycete fungus Penicillium digitatum. Genomics 2021, 113, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenführer, J. Gene set enrichment analysis with topGO. Bioconductor Improv. 2009, 27, 1–26. [Google Scholar]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Garrigues, S.; Manzanares, P.; Marcos, J.F. Application of recyclable CRISPR/Cas9 tools for targeted genome editing in the postharvest pathogenic fungi Penicillium digitatum and Penicillium expansum. Curr. Genet. 2022, 68, 515–529. [Google Scholar] [CrossRef]
- Poudel, R.; Rodriguez, L.T.; Reisch, C.R.; Rivers, A.R. GuideMaker: Software to design CRISPR-Cas guide RNA pools in non-model genomes. GigaScience 2022, 11, giac007. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yang, X.; Ruan, R.; Fu, H.; Li, H. Csn5 Is Required for the Conidiogenesis and Pathogenesis of the Alternaria alternata Tangerine Pathotype. Front. Microbiol. 2018, 9, 508. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, C.; Zhu, C.; Sun, X.; Ruan, R.; Li, H. Os2 MAP kinase-mediated osmostress tolerance in Penicillium digitatum is associated with its positive regulation on glycerol synthesis and negative regulation on ergosterol synthesis. Microbiol. Res. 2014, 169, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, J.; Liu, J.; Yuan, Y.; Li, N.; He, M.; Qi, T.; Hui, G.; Xiong, L.; Liu, D. Novel mutations in CYP51B from Penicillium digitatum involved in prochloraz resistance. J. Microbiol. 2014, 52, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, S.; Qin, T.; Li, N.; Niu, Y.; Li, D.; Yuan, Y.; Geng, H.; Xiong, L.; Liu, D. Whole transcriptome analysis of Penicillium digitatum strains treatmented with prochloraz reveals their drug-resistant mechanisms. BMC Genom. 2015, 16, 855. [Google Scholar] [CrossRef] [PubMed]
- Bolton, M.D.; Ebert, M.K.; Faino, L.; Rivera-Varas, V.; de Jonge, R.; Van de Peer, Y.; Thomma, B.P.H.J.; Secor, G.A. RNA-sequencing of Cercospora beticola DMI-sensitive and -resistant isolates after treatment with tetraconazole identifies common and contrasting pathway induction. Fungal Genet. Biol. 2016, 92, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Cao, Q.; Li, N.; Liu, D.; Yuan, Y. Transcriptome analysis of fungicide-responsive gene expression profiles in two Penicillium italicum strains with different response to the sterol demethylation inhibitor (DMI) fungicide prochloraz. BMC Genom. 2020, 21, 156. [Google Scholar] [CrossRef] [PubMed]
- Kwon, N.-J.; Garzia, A.; Espeso, E.A.; Ugalde, U.; Yu, J.-H. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol 2010, 77, 1203–1219. [Google Scholar] [CrossRef]
- Zhang, T.; Sun, X.; Xu, Q.; Candelas, L.G.; Li, H. The pH signaling transcription factor PacC is required for full virulence in Penicillium digitatum. Appl. Microbiol. Biotechnol. 2013, 97, 9087–9098. [Google Scholar] [CrossRef]
- Li, B.; Chen, Y.; Tian, S. Function of pH-dependent transcription factor PacC in regulating development, pathogenicity, and mycotoxin biosynthesis of phytopathogenic fungi. FEBS J. 2022, 289, 1723–1730. [Google Scholar] [CrossRef]
- Park, H.-S.; Yu, J.-H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 2012, 15, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Li, Z.; Wu, R.; Qin, Y.; Liu, G.; Qu, Y. Penicillium oxalicum PoFlbC regulates fungal asexual development and is important for cellulase gene expression. Fungal Genet. Biol. 2016, 86, 91–102. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xi, Y.; Zhang, J.; Fan, B.; Sun, M.; Cao, W.; Liu, X.; Gai, Y.; Shen, C.; Wang, H.; Wang, M. Transcriptome Analysis Reveals Potential Regulators of DMI Fungicide Resistance in the Citrus Postharvest Pathogen Penicillium digitatum. J. Fungi 2024, 10, 360. https://doi.org/10.3390/jof10050360
Xi Y, Zhang J, Fan B, Sun M, Cao W, Liu X, Gai Y, Shen C, Wang H, Wang M. Transcriptome Analysis Reveals Potential Regulators of DMI Fungicide Resistance in the Citrus Postharvest Pathogen Penicillium digitatum. Journal of Fungi. 2024; 10(5):360. https://doi.org/10.3390/jof10050360
Chicago/Turabian StyleXi, Yue, Jing Zhang, Botao Fan, Miaomiao Sun, Wenqian Cao, Xiaotian Liu, Yunpeng Gai, Chenjia Shen, Huizhong Wang, and Mingshuang Wang. 2024. "Transcriptome Analysis Reveals Potential Regulators of DMI Fungicide Resistance in the Citrus Postharvest Pathogen Penicillium digitatum" Journal of Fungi 10, no. 5: 360. https://doi.org/10.3390/jof10050360





