AP2XII-1 and AP2XI-2 Suppress Schizogony Gene Expression in Toxoplasma gondii
Abstract
:1. Introduction
2. Results
2.1. A Strong Correlation between the Downstream Promoters Bound by AP2XII-1 and AP2XI-2 and MORC
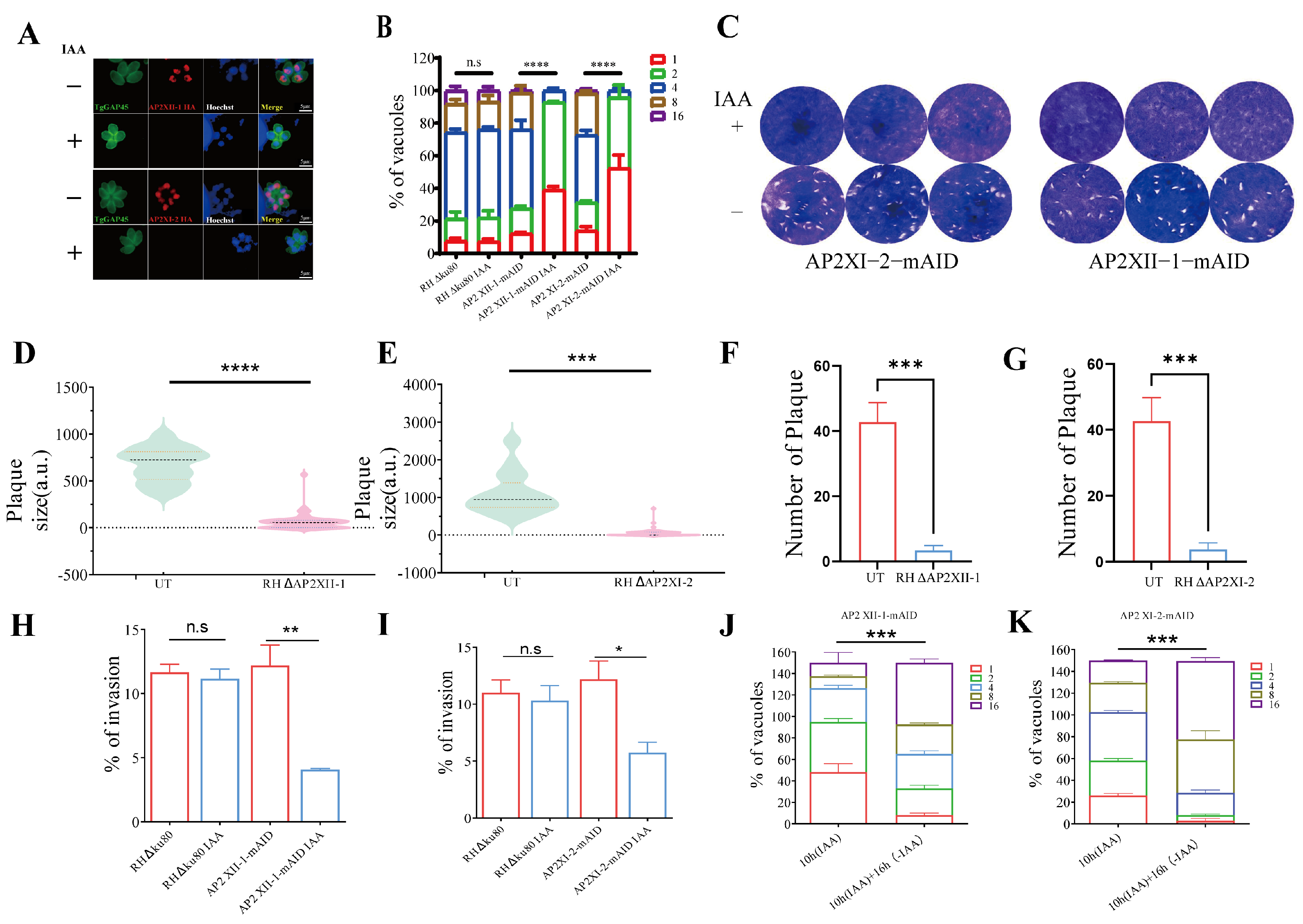
2.2. AP2XII-1 and AP2XI-2 Are Critical for Tachyzoite Replication In Vitro
2.3. The Transcriptional Levels of Downstream Genes Regulated by AP2XII-1 Were Correlated with MORC
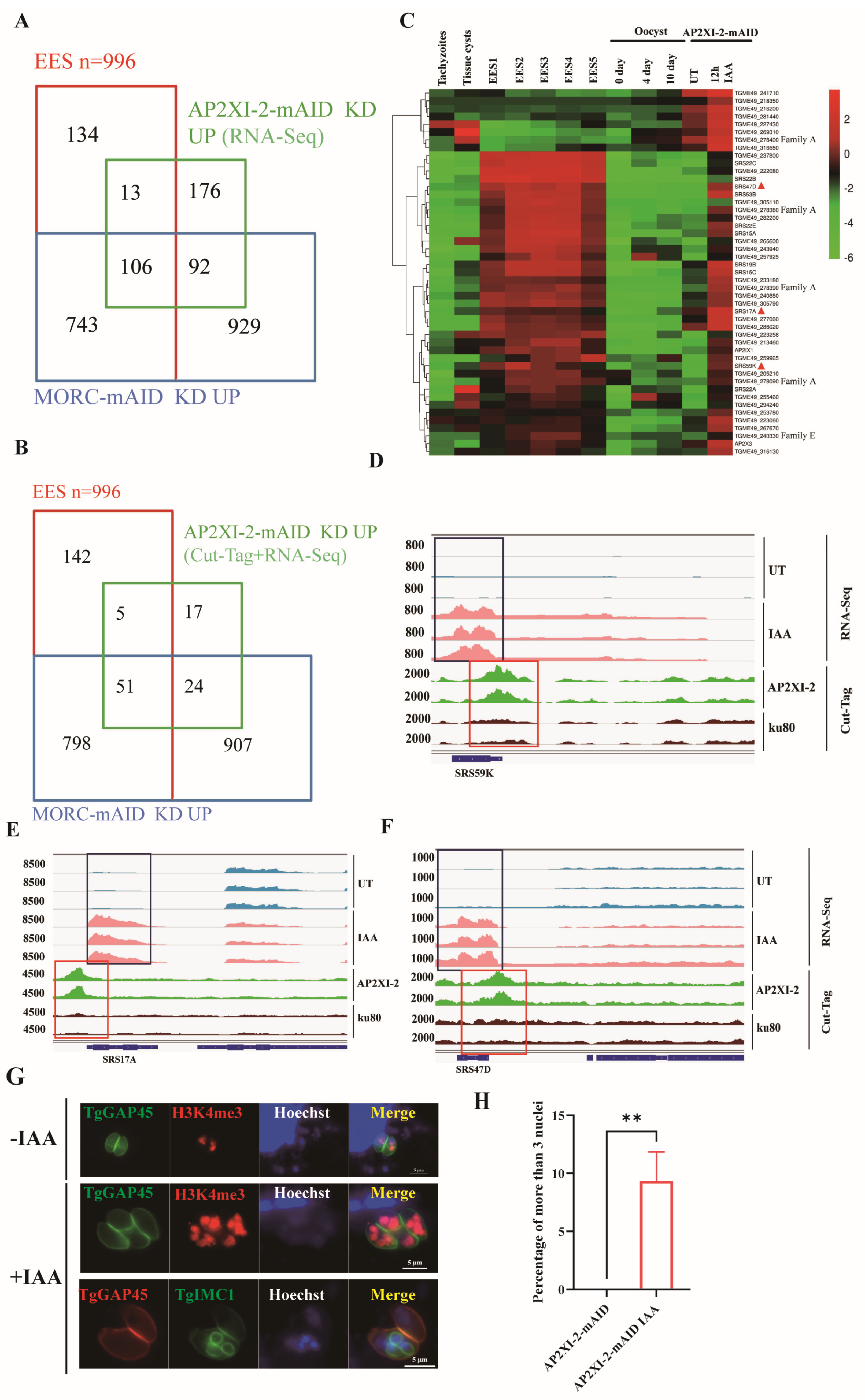
2.4. AP2XII-1 and AP2XI-2 Deficiency Induces the Expression of EES-Restricted Transcripts and Parasite Endopolygeny
2.5. AP2XII-1 and AP2XI-2 Co-Regulate Specific Genes in Enteroepithelial Stages
2.6. AP2XII-1 Is a Primary AP2 and Regulates the Expression of Multiple AP2 Genes
3. Discussion
4. Materials and Methods
4.1. Culture of Cells and Parasites
4.2. Generation of Transgenic T. gondii Strains
4.3. Intracellular Replication Assay and Invasion Assay
4.4. Plaque Assay
4.5. Immunoblotting and Immunofluorescence Assays
4.6. RNA-Seq and Data Analysis
4.7. Cut-Tag and Data Analysis
4.8. Luciferase Assays
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Montoya, J.G.; Liesenfeld, O. Toxoplasmosis. Lancet 2004, 363, 1965–1976. [Google Scholar] [CrossRef] [PubMed]
- Kochanowsky, J.A.; Koshy, A.A. Toxoplasma gondii. Curr. Biol. 2018, 28, R770–R771. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Parmley, S.F. Toxoplasma gondii expresses two distinct lactate dehydrogenase homologous genes during its life cycle in intermediate hosts. Gene 1997, 184, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Martorelli Di Genova, B.; Wilson, S.K.; Dubey, J.P.; Knoll, L.J. Intestinal delta-6-desaturase activity determines host range for Toxoplasma sexual reproduction. PLoS Biol. 2019, 17, e3000364. [Google Scholar] [CrossRef] [PubMed]
- Dubey, J.P.; Miller, N.L.; Frenkel, J.K. The Toxoplasma gondii oocyst from cat feces. J. Exp. Med. 1970, 132, 636–662. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Iyer, L.M.; Wellems, T.E.; Miller, L.H. Plasmodium biology: Genomic gleanings. Cell 2003, 115, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, W.J., Jr.; Hakimi, M.A. Histone mediated gene activation in Toxoplasma gondii. Mol. Biochem. Parasitol. 2006, 148, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Gissot, M.; Kelly, K.A.; Ajioka, J.W.; Greally, J.M.; Kim, K. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog. 2007, 3, e77. [Google Scholar] [CrossRef] [PubMed]
- Farhat, D.C.; Swale, C.; Dard, C.; Cannella, D.; Ortet, P.; Barakat, M.; Sindikubwabo, F.; Belmudes, L.; De Bock, P.J.; Couté, Y.; et al. A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment. Nat. Microbiol. 2020, 5, 570–583. [Google Scholar] [CrossRef]
- Gutterson, N.; Reuber, T.L. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 2004, 7, 465–471. [Google Scholar] [CrossRef]
- Xu, Z.S.; Chen, M.; Li, L.C.; Ma, Y.Z. Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 2011, 53, 570–585. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Xu, Z.S.; Huang, Y.; Tian, C.; Wang, F.; Xiong, A.S. Genome-wide analysis of AP2/ERF transcription factors in carrot (Daucus carota L.) reveals evolution and expression profiles under abiotic stress. Mol. Genet. Genom. 2015, 290, 2049–2061. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef] [PubMed]
- Antunes, A.V.; Shahinas, M.; Swale, C.; Farhat, D.C.; Ramakrishnan, C.; Bruley, C.; Cannella, D.; Robert, M.G.; Corrao, C.; Couté, Y.; et al. In vitro production of cat-restricted Toxoplasma pre-sexual stages. Nature 2024, 625, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Xue, L.; Yin, X.; Gupta, N.; Shen, B. AP2XII-1 is a negative regulator of merogony and presexual commitment in Toxoplasma gondii. mBio 2023, 14, e0178523. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.L.; Li, T.T.; Zhang, N.Z.; Wang, M.; Sun, L.X.; Zhang, Z.W.; Fu, B.Q.; Elsheikha, H.M.; Zhu, X.Q. The transcription factor AP2XI-2 is a key negative regulator of Toxoplasma gondii merogony. Nat. Commun. 2024, 15, 793. [Google Scholar] [CrossRef]
- Sidik, S.M.; Huet, D.; Ganesan, S.M.; Huynh, M.H.; Wang, T.; Nasamu, A.S.; Thiru, P.; Saeij, J.P.J.; Carruthers, V.B.; Niles, J.C.; et al. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell 2016, 166, 1423–1435.e12. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.M.; Sibley, L.D. Essential cGMP Signaling in Toxoplasma Is Initiated by a Hybrid P-Type ATPase-Guanylate Cyclase. Cell Host Microbe 2018, 24, 804–816.e6. [Google Scholar] [CrossRef]
- Wang, C.; Hu, D.; Tang, X.; Song, X.; Wang, S.; Zhang, S.; Duan, C.; Sun, P.; Suo, J.; Ma, H.; et al. Internal daughter formation of Toxoplasma gondii tachyzoites is coordinated by transcription factor TgAP2IX-5. Cell. Microbiol. 2021, 23, e13291. [Google Scholar] [CrossRef]
- Pittman, K.J.; Aliota, M.T.; Knoll, L.J. Dual transcriptional profiling of mice and Toxoplasma gondii during acute and chronic infection. BMC Genom. 2014, 15, 806. [Google Scholar] [CrossRef] [PubMed]
- Hehl, A.B.; Basso, W.U.; Lippuner, C.; Ramakrishnan, C.; Okoniewski, M.; Walker, R.A.; Grigg, M.E.; Smith, N.C.; Deplazes, P. Asexual expansion of Toxoplasma gondii merozoites is distinct from tachyzoites and entails expression of non-overlapping gene families to attach, invade, and replicate within feline enterocytes. BMC Genom. 2015, 16, 66. [Google Scholar] [CrossRef]
- Ramakrishnan, C.; Maier, S.; Walker, R.A.; Rehrauer, H.; Joekel, D.E.; Winiger, R.R.; Basso, W.U.; Grigg, M.E.; Hehl, A.B.; Deplazes, P.; et al. An experimental genetically attenuated live vaccine to prevent transmission of Toxoplasma gondii by cats. Sci. Rep. 2019, 9, 1474. [Google Scholar] [CrossRef] [PubMed]
- Fritz, H.M.; Buchholz, K.R.; Chen, X.; Durbin-Johnson, B.; Rocke, D.M.; Conrad, P.A.; Boothroyd, J.C. Transcriptomic analysis of toxoplasma development reveals many novel functions and structures specific to sporozoites and oocysts. PLoS ONE 2012, 7, e29998. [Google Scholar] [CrossRef]
- Kim, K.; Weiss, L.M. Toxoplasma gondii: The model apicomplexan. Int. J. Parasitol. 2004, 34, 423–432. [Google Scholar] [CrossRef]
- Dubey, J.P.; Lindsay, D.S.; Speer, C.A. Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin. Microbiol. Rev. 1998, 11, 267–299. [Google Scholar] [CrossRef]
- Radke, J.B.; Lucas, O.; De Silva, E.K.; Ma, Y.; Sullivan, W.J., Jr.; Weiss, L.M.; Llinas, M.; White, M.W. ApiAP2 transcription factor restricts development of the Toxoplasma tissue cyst. Proc. Natl. Acad. Sci. USA 2013, 110, 6871–6876. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, C.; Walker, R.A.; Eichenberger, R.M.; Hehl, A.B.; Smith, N.C. The merozoite-specific protein, TgGRA11B, identified as a component of the Toxoplasma gondii parasitophorous vacuole in a tachyzoite expression model. Int. J. Parasitol. 2017, 47, 597–600. [Google Scholar] [CrossRef]
- Hong, D.P.; Radke, J.B.; White, M.W. Opposing Transcriptional Mechanisms Regulate Toxoplasma Development. mSphere 2017, 2, e00347-16. [Google Scholar] [CrossRef]
- Waldman, B.S.; Schwarz, D.; Wadsworth, M.H., 2nd; Saeij, J.P.; Shalek, A.K.; Lourido, S. Identification of a Master Regulator of Differentiation in Toxoplasma. Cell 2020, 180, 359–372.e16. [Google Scholar] [CrossRef]
- Walker, R.; Gissot, M.; Croken, M.M.; Huot, L.; Hot, D.; Kim, K.; Tomavo, S. The Toxoplasma nuclear factor TgAP2XI-4 controls bradyzoite gene expression and cyst formation. Mol. Microbiol. 2013, 87, 641–655. [Google Scholar] [CrossRef]
- Lesage, K.M.; Huot, L.; Mouveaux, T.; Courjol, F.; Saliou, J.M.; Gissot, M. Cooperative binding of ApiAP2 transcription factors is crucial for the expression of virulence genes in Toxoplasma gondii. Nucleic Acids Res. 2018, 46, 6057–6068. [Google Scholar] [CrossRef]
- Xue, Y.; Theisen, T.C.; Rastogi, S.; Ferrel, A.; Quake, S.R.; Boothroyd, J.C. A single-parasite transcriptional atlas of Toxoplasma Gondii reveals novel control of antigen expression. eLife 2020, 9, e54129. [Google Scholar] [CrossRef]
- Khelifa, A.S.; Guillen Sanchez, C.; Lesage, K.M.; Huot, L.; Mouveaux, T.; Pericard, P.; Barois, N.; Touzet, H.; Marot, G.; Roger, E.; et al. TgAP2IX-5 is a key transcriptional regulator of the asexual cell cycle division in Toxoplasma gondii. Nat. Commun. 2021, 12, 116. [Google Scholar] [CrossRef]
- Farhat, D.C.; Hakimi, M.A. The developmental trajectories of Toxoplasma stem from an elaborate epigenetic rewiring. Trends Parasitol. 2022, 38, 37–53. [Google Scholar] [CrossRef]
- Jeninga, M.D.; Quinn, J.E.; Petter, M. ApiAP2 Transcription Factors in Apicomplexan Parasites. Pathogens 2019, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Behnke, M.S.; Wootton, J.C.; Lehmann, M.M.; Radke, J.B.; Lucas, O.; Nawas, J.; Sibley, L.D.; White, M.W. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS ONE 2010, 5, e12354. [Google Scholar] [CrossRef] [PubMed]
- Jeffers, V.; Tampaki, Z.; Kim, K.; Sullivan, W.J., Jr. A latent ability to persist: Differentiation in Toxoplasma gondii. Cell. Mol. Life Sci. 2018, 75, 2355–2373. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Cui, X.; Fan, S.; Liu, J.; Zhang, X.; Wu, Y.; Liu, Q. Comprehensive Characterization of Toxoplasma Acyl Coenzyme A-Binding Protein TgACBP2 and Its Critical Role in Parasite Cardiolipin Metabolism. mBio 2018, 9, e01597-18. [Google Scholar] [CrossRef]
- Brown, K.M.; Long, S.; Sibley, L.D. Plasma Membrane Association by N-Acylation Governs PKG Function in Toxoplasma gondii. mBio 2017, 8, e00375-17. [Google Scholar] [CrossRef]
- Shen, B.; Sibley, L.D. Toxoplasma aldolase is required for metabolism but dispensable for host-cell invasion. Proc. Natl. Acad. Sci. USA 2014, 111, 3567–3572. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yang, M.; Yu, H.; Yu, X.; Liang, S.; Hu, Y.; Luo, Y.; Izsvák, Z.; Sun, C.; Wang, J. Chemical-induced chromatin remodeling reprograms mouse ESCs to totipotent-like stem cells. Cell Stem Cell 2022, 29, 400–418.e13. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef] [PubMed]
- Yashar, W.M.; Kong, G.; VanCampen, J.; Curtiss, B.M.; Coleman, D.J.; Carbone, L.; Yardimci, G.G.; Maxson, J.E.; Braun, T.P. GoPeaks: Histone modification peak calling for CUT & Tag. Genome Biol. 2022, 23, 144. [Google Scholar]
- Singh, R.; Zhang, F.; Li, Q. Assessing reproducibility of high-throughput experiments in the case of missing data. Stat. Med. 2022, 41, 1884–1899. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.H.; Brown, J.B.; Huang, H.Y.; Bickel, P.J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 2011, 5, 1752–1779. [Google Scholar] [CrossRef]
- Srivastava, S.; Holmes, M.J.; White, M.W.; Sullivan, W.J., Jr. Toxoplasma gondii AP2XII-2 Contributes to Transcriptional Repression for Sexual Commitment. mSphere 2023, 8, e0060622. [Google Scholar] [CrossRef]
- Huang, S.; Holmes, M.J.; Radke, J.B.; Hong, D.P.; Liu, T.K.; White, M.W.; Sullivan, W.J., Jr. Toxoplasma gondii AP2IX-4 Regulates Gene Expression during Bradyzoite Development. mSphere 2017, 2, e00054-17. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Y.; Shi, Y.; Xue, Y.; Hu, D.; Song, X. AP2XII-1 and AP2XI-2 Suppress Schizogony Gene Expression in Toxoplasma gondii. Int. J. Mol. Sci. 2024, 25, 5527. https://doi.org/10.3390/ijms25105527
Jiang Y, Shi Y, Xue Y, Hu D, Song X. AP2XII-1 and AP2XI-2 Suppress Schizogony Gene Expression in Toxoplasma gondii. International Journal of Molecular Sciences. 2024; 25(10):5527. https://doi.org/10.3390/ijms25105527
Chicago/Turabian StyleJiang, Yucong, Yuehong Shi, Yingying Xue, Dandan Hu, and Xingju Song. 2024. "AP2XII-1 and AP2XI-2 Suppress Schizogony Gene Expression in Toxoplasma gondii" International Journal of Molecular Sciences 25, no. 10: 5527. https://doi.org/10.3390/ijms25105527





