The Development of a One-Step RT-qPCR for the Detection and Quantification of Viable Forms of Trypanosoma cruzi in Açai Samples from Areas at Risk of Chagas Disease through Oral Transmission
Abstract
:1. Introduction
2. Results
2.1. The Establishment of the RNA Exogenous Internal Positive Control
2.2. Analytical Sensitivity for Viable T. cruzi Detection
2.3. The Reportable Range of Viable T. cruzi Detection in the One-Step RT-qPCR Assay (Linearity Assay)
2.4. Detection of Viable T. cruzi from Different DTUs by One-Step RT-qPCR (Inclusivity Assay)
2.5. Precision on the Detection of T. cruzi RNA by One-Step RT-qPCR
2.6. The Validation of the One-Step RT-qPCR Assay with Açai Samples from the Field
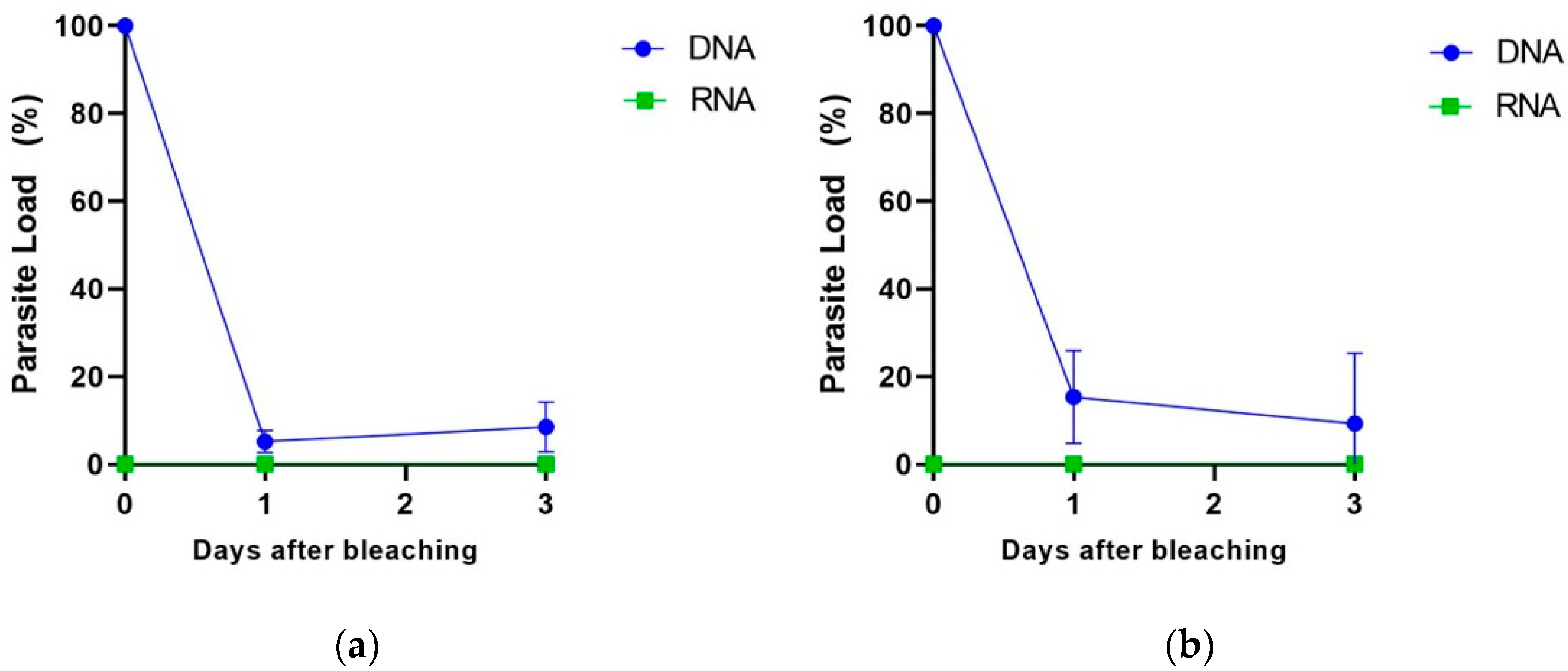
2.7. Quantification of T. cruzi DNA and RNA in Bleached Açai Samples
3. Discussion
4. Materials and Methods
4.1. Açaí Samples
4.2. Trypanosoma cruzi Cultivation
4.3. Preparation of Guanidine-EDTA Açaí (GEA) Samples
4.4. DNA and RNA Extraction
4.5. One Step RT-qPCR for T. cruzi Detection and Quantification
4.6. Inclusivity Assay
4.7. Sensitivity Assay
4.8. Standard Curves Preparation
4.9. Validation with Açaí Pulp Samples from Municipality of Coari (AM)
4.10. Açaí Fruit Bleaching
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moncayo, A.; Silveira, A.C. Current epidemiological trends for Chagas disease in Latin America and future challenges in epidemiology, surveillance and health policy. Memórias Do Inst. Oswaldo Cruz. 2009, 104, 17–30. [Google Scholar] [CrossRef]
- Coura, J.R. Chagas disease: What is known and what is needed—A 91 background article. Memórias Do Inst. Oswaldo Cruz. 2007, 102, 113–122. [Google Scholar] [CrossRef]
- Pérez-Molina, J.A.; Molina, I. Chagas disease. Lancet 2018, 391, 82–94. [Google Scholar] [CrossRef]
- Alarcón de Noya, B.; Jackson, Y. Chagas disease epidemiology: From Latin America to the world. In Chagas Disease: A Neglected Tropical Disease; Springer: Berlin/Heidelberg, Germany, 2020; pp. 27–36. [Google Scholar]
- World Health Organization. Neglected Tropical Diseases (NTDs). Available online: https://www.who.int/health-topics/neglected-tropical-diseases#tab=tab_1 (accessed on 6 December 2021).
- Dias, J.C. Notas sobre o Trypanosoma cruzi e suas características bio-ecológicas, como agente de enfermidades transmitidas por alimentos. Rev. Da Soc. Bras. De Med. Trop. 2006, 39, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Nóbrega, A.A.; Garcia, M.H.; Tatto, E.; Obara, M.T.; Costa, E.; Sobel, J.; Araujo, W.N. Oral transmission of chagas disease by consumption of Açaí palm fruit, Brazil. Emerg. Infect. Dis. 2009, 15, 653–655. [Google Scholar] [CrossRef]
- Fregonesi, B.M.; Yokosawa, C.E.; Okada, I.A.; Massafera, G.; Costa, T.M.B.; Prado, S.P.T. Frozen açaí pulp: Nutritional, physicochemical, microscopic characteristics and labeling evaluation. Inst. Adolfo Lutz 2010, 69, 387–395. [Google Scholar] [CrossRef]
- Mattos, E.C.C.; Meira-Strejevitch, C.S.; Marciano, M.A.M.; Faccini, C.C.; Lourenço, A.M.; Pereira-Chioccola, V.L. Molecular detection of Trypanosoma cruzi in acai pulp and sugarcane juice. Acta Trop. 2017, 176, 311–315. [Google Scholar] [CrossRef]
- Dias, J.C.P.P.; Neto, V.A.; de Albuquerque Luna, E.J. Alternative 93 transmission mechanisms of Trypanosoma cruzi in Brazil and proposals for their prevention. Rev. Da Soc. Bras. De Med. Trop. 2011, 44, 375–379. [Google Scholar] [CrossRef]
- Coura, J.R.; Viñas, P.A.; Junqueira, A.C.V. Ecoepidemiology, Short history and control of chagas disease in the endemic countries and the new challenge for non-endemic countries. Memórias Do Inst. Oswaldo Cruz. 2014, 109, 856–862. [Google Scholar] [CrossRef]
- Ferreira, R.T.B.; Branquinho, M.R.; Leite, P.C. Oral transmission of Chagas disease by consumption of açaí: A challenge for Health Surveillance. Vig Sanit. Debate 2014, 2, 4–11. [Google Scholar] [CrossRef]
- Shikanai-Yasuda, M.A.; Carvalho, N.B. Oral transmission of Chagas disease. Clin. Infect. Dis. 2012, 54, 845–852. [Google Scholar] [CrossRef]
- Monteiro, S. Açaí: Da fruta exótica à vedete de consumo. Frutas E Deriv. Epagri-Sede Camp. 2006, 1, 29–32. [Google Scholar]
- Rogez, H.L.G.; Aguiar, F.S. Contaminação da Bebida do Açaí Envolvendo o Trypanosoma cruzi. In Tecnologias para Inovação nas Cadeias Euterpe; Pessoa, J.D.C., Teixeira, G.H.A., Eds.; Embrapa: Brasília, Brazil, 2012; pp. 205–228. [Google Scholar]
- Bezerra, V.S. Açaí seguro: Choque térmico nos frutos de açaí como recomendação para eliminação do agente causador da doença de Chagas. In Embrapa Amapá-Nota Técnica/Nota; Embrapa: Macapá, Brasil, 2018. [Google Scholar]
- Passos, L.A.C.; Guaraldo, A.M.A.; Barbosa, R.L.; Dias, V.L.; Pereira, K.S.; Schmidt, F.L.; Franco, R.M.B.; Alves, D.P. Sobrevivência e infectividade do Trypanosoma cruzi na polpa de açaí: Estudo in vitro e in vivo. Epidemiol. E Serviços Saúde 2012, 21, 223–232. [Google Scholar] [CrossRef]
- Ferreira, R.T.B.; Cabral, M.L.; Martins, R.S.; Araujo, P.F.; da Silva, S.A.; Britto, C.; Branquinho, M.R.; Cardarelli-Leite, P.; Moreira, O.C. Detection and genotyping of Trypanosoma cruzi in açaí products sold in Rio de Janeiro and Pará, Brazil. Parasites Vectors 2018, 11, 233. [Google Scholar] [CrossRef] [PubMed]
- Finamore-Araujo, P.; Faier-Pereira, A.; Brito, C.R.D.N.; Peres, E.G.; Yamaguchi, K.K.d.L.; Ferreira, R.T.B.; Moreira, O.C. Validation of a novel multiplex real-time PCR assay for Trypanosoma cruzi detection and quantification in açai pulp. PLoS ONE 2021, 16, e0246435. [Google Scholar] [CrossRef]
- Godoi, P.A.d.S.; Piechnik, C.A.; de Oliveira, A.C.; Sfeir, M.Z.; de Souza, E.M.; Rogez, H.; Soccol, V.T. qPCR for the detection of foodborne Trypanosoma cruzi. Parasitol. Int. 2017, 66, 563–566. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, A.C.; Soccol, V.T.; Rogez, H. Prevention methods of foodborne Chagas disease: Disinfection, heat treatment and quality control by RT-PCR. Int. J. Food Microbiol. 2019, 301, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, G.V.F.; Lima, J.S.; de Oliveira, A.C.S.; da Silva, J.B.; Roos, T.B.; de Moraes, C.M. SYBR Green qPCR Technique for Detection of Trypanosoma cruzi in Açaí Pulp. Foodborne Pathog. Dis. 2020, 17, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.T.B.; Melandre, A.M.; Cabral, M.L.; Branquinho, M.R.; Cardarelli-Leite, P. Extraction of Trypanosoma cruzi DNA from food: A contribution to the elucidation of acute Chagas disease outbreaks. Rev. Da Soc. Bras. De Med. Trop. 2016, 49, 190–195. [Google Scholar] [CrossRef]
- Masters, C.I.; Shallcross, J.A.; Mackey, B.M. Effect of stress treatments on detection of enterotoxigenic Listeria monocytogenes and Escherichia coli by polymerase chain reaction. J. Appl. Microbiol. 1994, 77, 73–79. [Google Scholar] [CrossRef]
- Keer, J.T.; Birch, L. Molecular methods for the assessment of bacterial viability. J. Microbiol. Methods 2003, 53, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tarleton, R.L. Parasite persistence correlates with disease severity and localization in chronic Chagas’ disease. J. Infect. Dis. 1999, 180, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, G.E.C.; Masters, C.I.; Shallcross, J.A.; Mackey, B.M. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl. Environ. Microbiol. 1998, 64, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- van der Meide, W.F.; Schoone, G.J.; Faber, W.R.; Zeegelaar, J.E.; de Vries, H.J.C.; Özbel, Y.; Fat, R.F.M.L.A.; Coelho, L.I.A.R.C.; Kassi, M.; Schallig, H.D.F.H. Quantitative nucleic acid sequence-based assay as a new molecular tool for detection and quantification of Leishmania parasites in skin biopsy samples. J. Clin. Microbiol. 2005, 43, 5560–5566. [Google Scholar] [CrossRef] [PubMed]
- Payne, R.; Wang, F.; Su, N.; Krell, J.; Zebrowski, A.; Yagüe, E.; Ma, X.-J.; Luo, Y.; Coombes, R.C. Viable circulating tumour cell detection using multiplex RNA in situ hybridisation predicts progression-free survival in metastatic breast cancer patients. Br. J. Cancer 2012, 106, 1790–1797. [Google Scholar] [CrossRef]
- Hellyer, T.J.; DesJardin, L.E.; Hehman, G.L.; Cave, M.D. Quantitative Analysis of mRNA as a Marker for Viability of Mycobacterium tuberculosis. Eisenach J. Clin. Microbiol. 1999, 37, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Finamore-Araujo, P.; Silva da Fonseca, G.L.; Vieira, C.S.; de Castro, D.P.; Moreira, O.C. RNA as a feasible marker of Trypanosoma cruzi viability during the parasite interaction with the triatomine vector Rhodnius prolixus (Hemiptera, Triatominae). PLoS Negl. Trop. Dis. 2022, 16, e0010535. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, A.C.; Thomaz-Soccol, V.; Tadra-Sfeir, M.Z.; de Souza, E.M.; Crespo-Lopez, M.E.; Rogez, H. Preventing Chagas disease: A new RT-qPCR method for rapid and specific quantification of viable Trypanosoma cruzi for food safety. Int. J. Food Microbiol. 2021, 144, 110368. [Google Scholar] [CrossRef]
- Barbosa, R.L.; Dias, V.L.; Lorosa, E.S.; Costa, E.d.G.; Pereira, K.S.; Gilioli, R.; Guaraldo, A.M.A.; Passos, L.A.C. Virulence of Trypanosoma cruzi from vector and reservoir in in natura açai pulp resulting in foodborne acute Chagas disease at Pará State, Brazil. Exp. Parasitol. 2019, 197, 68–75. [Google Scholar] [CrossRef]
- Costa, A.D.; Jacomasso, T.; Mattos, E.C.; Farias, A.B.; Rampazzo, R.C.; Pinto, R.S.; Tassi, W.; Marciano, M.A.M.; Perei-ra-Chioccola, V.L.; Murphy, H.R.; et al. Ready-to-use qPCR for detection of Cyclospora cayetanensis or Trypanosoma cruzi in food matrices. Food Waterborne Parasitol. 2021, 22, e00111. [Google Scholar] [CrossRef]
- Johnson, D.R.; Lee, P.K.; Holmes, V.F.; Alvarez-Cohen, L. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol. 2005, 71, 3866–3871. [Google Scholar] [CrossRef] [PubMed]
- Avila, H.A.; Sigman, D.S.; Cohen, L.M.; Millikan, R.C.; Simpson, L. Polymerase chain reaction amplification of Trypanosoma cruzi kinetoplast minicircle DNA isolated from whole blood lysates: Diagnosis of chronic Chagas disease. Mol. Biochem. Parasitol. 1991, 48, 211–221. [Google Scholar] [CrossRef]
- Bowtell, D.D.L. Rapid isolation of eukaryotic DNA. Anal. Biochem. 1987, 162, 463–465. [Google Scholar] [CrossRef] [PubMed]
- Britto, C.; Cardoso, M.A.; Wincker, P.; Morel, C.M. A simple protocol for the physical cleavage of Trypanosoma cruzi kinetoplast DNA present in blood samples and its use in polymerase chain reaction (PCR)-based diagnosis of chronic Chagas disease. Mem. Inst. Oswaldo Cruz. 1993, 88, 171–172. [Google Scholar] [CrossRef] [PubMed]
- Moreira, O.C.; Ramírez, J.D.; Velázquez, E.; Melo, M.F.; Lima-Ferreira, C.; Guhl, F.; Sosa-Estani, S.; Marin-Neto, J.A.; Morillo, C.A.; Britto, C. Towards the establishment of a consensus real-time PCR to monitor Trypanosoma cruzi parasitemia in patients with chronic Chagas disease cardiomyopathy: A substudy from the Benefit trial. Acta Trop. 2013, 125, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Neves, V.C. Avaliação de Protocolos de Extração de DNA Empregados na Detecção de Trypanosoma cruzi Chagas, 1909 por PCR em Triatomíneos. Ph.D. Thesis, Fundação Oswaldo Cruz, Rio de Janeiro, Brasil, 2010. [Google Scholar]
- Duffy, T.; Cura, C.I.; Ramirez, J.C.; Abate, T.; Cayo, N.M.; Parrado, R.; Bello, Z.D.; Velazquez, E.; Muñoz-Calderon, A.; Juiz, N.A.; et al. Analytical performance of a multiplex Real-Time PCR assay using TaqMan probes for quantification of Trypanosoma cruzi satellite DNA in blood samples. PLoS Negl. Trop. Dis. 2013, 7, e2000. [Google Scholar] [CrossRef]
- Ramírez, J.C.; Cura, C.I.; Moreira, O.d.C.; Lages-Silva, E.; Juiz, N.; Velázquez, E.; Alberti, A.; Pavia, P.; Flores-Chávez, M.D.; Muñoz-Calderón, A.; et al. Analytical validation of quantitative real-time PCR methods for quantification of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. J. Mol. Diagn. 2015, 17, 605–615. [Google Scholar] [CrossRef]
- Clayton, C. Regulation of gene expression in trypanosomatids: Living with polycistronic transcription. Open Biol. 2019, 9, 190072. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Teixeira, S.M. Control of gene expression in Trypanosomatidae. Braz. J. Med. Biol. Res. 1998, 12, 1503–1516. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Calvillo, S.; Vizuet-de-Rueda, J.C.; Florencio-Martínez, L.E.; Manning-Cela, R.G.; Figueroa-Angulo, E.E. Gene expression in trypanosomatid parasites. J. Biomed. Biotechnol. 2010, 2010, 525241. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pinheiro, E.T.; Candeiro, G.T.; Teixeira, S.R.; Shin, R.C.; Prado, L.C.; Gavini, G.; Mayer, M.P.A. RNA-based Assay Demonstrated Enterococcus faecalis 59 Metabolic Activity after Chemomechanical Procedures. J. Endod. 2015, 41, 1441–1444. [Google Scholar] [CrossRef]
- Norouzi, R.; Dalimi, A.; Moghadam, M.F.F. Comparison of a Nucleic Acid Sequence-based Amplification (NASBA) and real-time reverse transcriptase PCR methods for detection of Toxoplasma gondii in rat blood samples. J. Zoonotic Dis. 2016, 1, 15–23. [Google Scholar]





| T. cruzi (Par. Eq/mL) | T. cruzi Ct Mean ± SD | RNA-IAC Ct Mean ± SD |
|---|---|---|
| 5 | 28.45 ± 0.38 | 22.52 ± 0.06 |
| 1 | 32.39 ± 0.13 | 23.27 ± 0.01 |
| 0.5 | 32.31 ± 0.15 | 23.29 ± 0.10 |
| 0.1 | 34.27 ± 0.01 | 23.54 ± 0.28 |
| Concentration (Par. Eq/mL) | Tc I | Tc III | Tc IV | |||
|---|---|---|---|---|---|---|
| Ct Mean ± SD | Ct Mean ± SD | Ct Mean ± SD | ||||
| T. cruzi | RNA-IAC | T. cruzi | RNA-IAC | T. cruzi | RNA-IAC | |
| 104 | 20.83 ± 0.98 | 22.98 ± 0.18 | 23.69 ± 1.74 | 24.74 ± 0.28 | 23.43 ± 0.56 | 23.73 ± 0.50 |
| 103 | 24.58 ± 0.25 | 22.40 ± 0.20 | 26.62 ± 0.23 | 27.14 ± 0.35 | 27.23 ± 0.49 | 24.86 ± 0.80 |
| 102 | 26.43 ± 0.47 | 22.67 ± 0.25 | 30.90 ± 1.99 | 25.00 ± 0.70 | 31.78 ± 0.49 | 25.45 ± 0.17 |
| 10 | 26.74 ± 0.12 | 22.55 ± 0.12 | 34.82 ± 0.84 | 26.05 ± 0.0 | 32.84 ± 1.24 | 25.74 ± 0.98 |
| 1 | 28.49 ± 0.92 | 23.80 ± 0.34 | NA 1 | 25.12 ± 0.44 | 34.51 ± 1.63 | 25.55 ± 0.4 |
| 0.1 | 29.36 ± 0.51 | 23.53 ± 0.67 | NA 1 | 26.85 ± 0.05 | 40.08 ± 4.31 | 26.89 ± 0.35 |
| Sample Concentration | 2 Par. Eq/mL | 1 Par. Eq/mL | 0.5 Par. Eq/mL |
|---|---|---|---|
| Positive results | 100% | 100% | 97.50% |
| Ct mean (±SD) | 30.91 (0.54) | 32.07 (0.48) | 32.47 (0.65) |
| Coefficient of variation | 1.74% | 1.49% | 2.01% |
| Target | Primers/Probes | Sequences | Amplicon size | Reference |
|---|---|---|---|---|
| T. cruzi (FAM/NFQ-MGB) | Cruzi 1 (Forward) | 5′-AAT CGG CTG ATC GTT TTC GA-3′ | 165 bp | Duffy et al., 2013 [41] |
| Cruzi 2 (Reverse) | 5′-AAT TCC TCC AAG CAG CGG ATA-3′ | |||
| Cruzi 3 (Probe) | 5′-CAC ACA CTG GAC ACC AA-3′ | |||
| exo-IPC (VIC/TAMRA) | Not Available | Not Available | Not Available | Applied Biosystems (Cat. Number: 4308323), Finamore-Araujo et al., 2021 [19] |
| RNA-IAC (VIC/NFQ-MGB) | Forward | 5′-TAC AAC ACC CCA ACA TCT TCG A-3′ | Not Available | Promega (Cat. Number: L4561), Johnson et al., 2005 [35] |
| Reverse | 5′-GGA AGT TCA CCG GCG TCA T-3′ | |||
| Probe | 5′-CGG GCG TGG CAG GTC TTC CC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faier-Pereira, A.; Finamore-Araujo, P.; Brito, C.R.d.N.; Peres, E.G.; de Lima Yamaguchi, K.K.; de Castro, D.P.; Moreira, O.C. The Development of a One-Step RT-qPCR for the Detection and Quantification of Viable Forms of Trypanosoma cruzi in Açai Samples from Areas at Risk of Chagas Disease through Oral Transmission. Int. J. Mol. Sci. 2024, 25, 5531. https://doi.org/10.3390/ijms25105531
Faier-Pereira A, Finamore-Araujo P, Brito CRdN, Peres EG, de Lima Yamaguchi KK, de Castro DP, Moreira OC. The Development of a One-Step RT-qPCR for the Detection and Quantification of Viable Forms of Trypanosoma cruzi in Açai Samples from Areas at Risk of Chagas Disease through Oral Transmission. International Journal of Molecular Sciences. 2024; 25(10):5531. https://doi.org/10.3390/ijms25105531
Chicago/Turabian StyleFaier-Pereira, Amanda, Paula Finamore-Araujo, Carlos Ramon do Nascimento Brito, Eldrinei Gomes Peres, Klenicy Kazumy de Lima Yamaguchi, Daniele Pereira de Castro, and Otacilio C. Moreira. 2024. "The Development of a One-Step RT-qPCR for the Detection and Quantification of Viable Forms of Trypanosoma cruzi in Açai Samples from Areas at Risk of Chagas Disease through Oral Transmission" International Journal of Molecular Sciences 25, no. 10: 5531. https://doi.org/10.3390/ijms25105531






