Effect of Obstructive Sleep Apnea during Pregnancy on Fetal Development: Gene Expression Profile of Cord Blood
Abstract
:1. Introduction
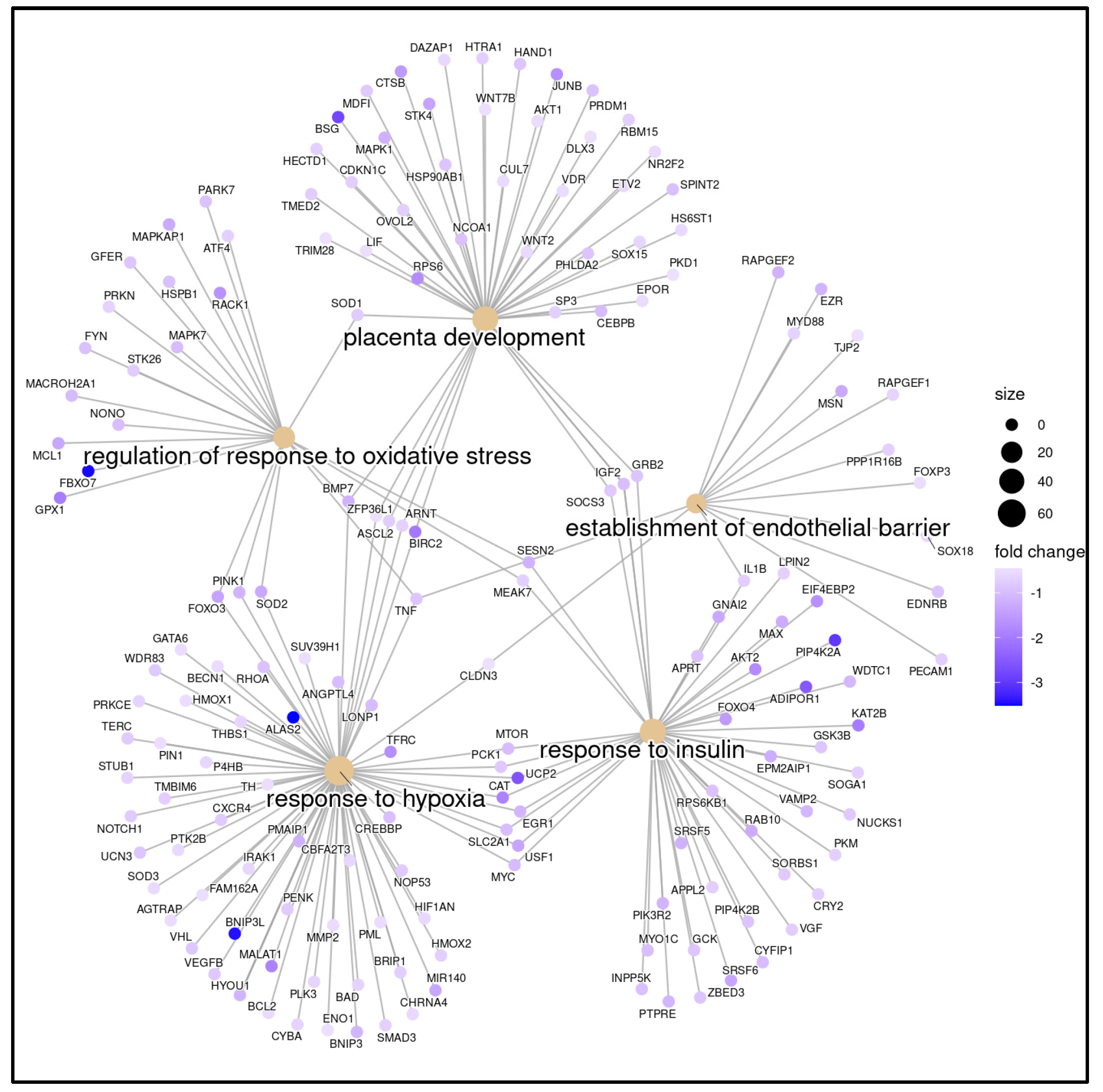
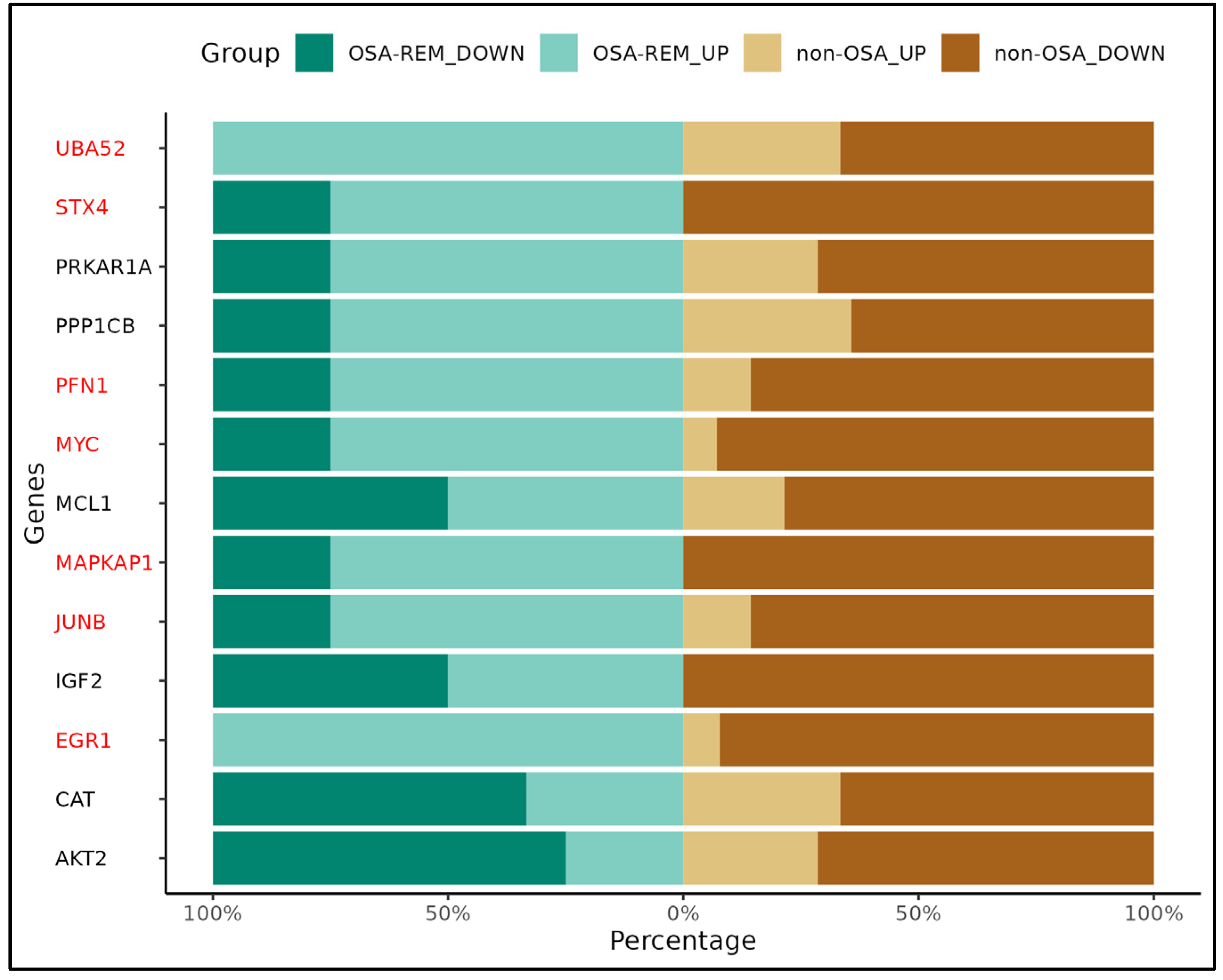
2. Results
2.1. Subjects of Study
2.2. Gene Expression Profiling Analysis
2.3. Gene Validation (qRT-PCR)
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Clinical and Sleep Evaluation
4.3. Sample Acquisition and Preparation
4.4. Gene Expression Analysis
4.5. qRTPCR Validation
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peppard, P.E.; Young, T.; Barnet, J.H.; Palta, M.; Hagen, E.W.; Hla, K.M. Increased Prevalence of Sleep-Disordered Breathing in Adults. Am. J. Epidemiol. 2013, 177, 1006–1014. [Google Scholar] [CrossRef]
- Pamidi, S.; Kimoff, R.J. Maternal Sleep-Disordered Breathing. Chest 2018, 153, 1052–1066. [Google Scholar] [CrossRef] [PubMed]
- Izci, B.; Riha, R.L.; Martin, S.E.; Vennelle, M.; Liston, W.A.; Dundas, K.C.; Calder, A.A.; Douglas, N.J. The Upper Airway in Pregnancy and Pre-Eclampsia. Am. J. Respir. Crit. Care Med. 2003, 167, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Fernández, A.; Ribot Quetglas, C.; Herranz Mochales, A.; Álvarez Ruiz De Larrinaga, A.; Sánchez Barón, A.; Rodríguez Rodríguez, P.; Gil Gómez, A.V.; Pía Martínez, C.; Cubero Marín, J.P.; Barceló Nicolau, M.; et al. Influence of Obstructive Sleep Apnea on Systemic Inflammation in Pregnancy. Front. Med. 2021, 8, 674997. [Google Scholar] [CrossRef] [PubMed]
- Tayade, S.; Toshniwal, S. Obstructive Sleep Apnea in Pregnancy: A Narrative Review. Cureus 2022, 14, e30387. [Google Scholar] [CrossRef] [PubMed]
- Facco, F.L.; Parker, C.B.; Reddy, U.M.; Silver, R.M.; Koch, M.A.; Louis, J.M.; Basner, R.C.; Chung, J.H.; Nhan-Chang, C.-L.; Pien, G.W.; et al. Association Between Sleep-Disordered Breathing and Hypertensive Disorders of Pregnancy and Gestational Diabetes Mellitus. Obstet. Gynecol. 2017, 129, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Truong, K.K.; Guilleminault, C. Sleep Disordered Breathing in Pregnant Women: Maternal and Fetal Risk, Treatment Considerations, and Future Perspectives. Expert Rev. Respir. Med. 2018, 12, 177–189. [Google Scholar] [CrossRef]
- Fung, A.M.; Wilson, D.L.; Lappas, M.; Howard, M.; Barnes, M.; O’Donoghue, F.; Tong, S.; Esdale, H.; Fleming, G.; Walker, S.P. Effects of Maternal Obstructive Sleep Apnoea on Fetal Growth: A Prospective Cohort Study. PLoS ONE 2013, 8, e68057. [Google Scholar] [CrossRef]
- Pamidi, S.; Marc, I.; Simoneau, G.; Lavigne, L.; Olha, A.; Benedetti, A.; Sériès, F.; Fraser, W.; Audibert, F.; Bujold, E.; et al. Maternal Sleep-Disordered Breathing and the Risk of Delivering Small for Gestational Age Infants: A Prospective Cohort Study. Thorax 2016, 71, 719–725. [Google Scholar] [CrossRef]
- Kneitel, A.W.; Treadwell, M.C.; O’Brien, L.M. Effects of Maternal Obstructive Sleep Apnea on Fetal Growth: A Case-Control Study. J. Perinatol. 2018, 38, 982–988. [Google Scholar] [CrossRef]
- Bourjeily, G.; Danilack, V.A.; Bublitz, M.H.; Lipkind, H.; Muri, J.; Caldwell, D.; Tong, I.; Rosene-Montella, K. Obstructive Sleep Apnea in Pregnancy Is Associated with Adverse Maternal Outcomes: A National Cohort. Sleep Med. 2017, 38, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Zhang, X.; Wang, Y.; Li, J.; Xu, Y.; Song, X.; Su, S.; Zhu, X.; Vitiello, M.V.; Shi, J.; et al. Sleep Disturbances during Pregnancy and Adverse Maternal and Fetal Outcomes: A Systematic Review and Meta-Analysis. Sleep Med. Rev. 2021, 58, 101436. [Google Scholar] [CrossRef] [PubMed]
- Marshall, N.E.; Abrams, B.; Barbour, L.A.; Catalano, P.; Christian, P.; Friedman, J.E.; Hay, W.W.; Hernandez, T.L.; Krebs, N.F.; Oken, E.; et al. The Importance of Nutrition in Pregnancy and Lactation: Lifelong Consequences. Am. J. Obstet. Gynecol. 2022, 226, 607–632. [Google Scholar] [CrossRef]
- Ghesquière, L.; Deruelle, P.; Ramdane, Y.; Garabedian, C.; Charley-Monaca, C.; Dalmas, A.F. Obstructive Sleep Apnea in Obese Pregnant Women: A Prospective Study. PLoS ONE 2020, 15, e0238733. [Google Scholar] [CrossRef]
- Wilson, D.L.; Howard, M.E.; Fung, A.M.; O’Donoghue, F.J.; Barnes, M.; Lappas, M.; Walker, S.P. Sleep-Disordered Breathing Does Not Impact Maternal Outcomes in Women with Hypertensive Disorders of Pregnancy. PLoS ONE 2020, 15, e0232287. [Google Scholar] [CrossRef]
- Chami, H.A.; Gottlieb, D.J.; Redline, S.; Punjabi, N.M. Association between Glucose Metabolism and Sleep-Disordered Breathing during REM Sleep. Am. J. Respir. Crit. Care Med. 2015, 192, 1118–1126. [Google Scholar] [CrossRef]
- BaHammam, A.S.; Pirzada, A.R.; Pandi-Perumal, S.R. Neurocognitive, Mood Changes, and Sleepiness in Patients with REM-Predominant Obstructive Sleep Apnea. Sleep Breath. 2023, 27, 57–66. [Google Scholar] [CrossRef]
- Mokhlesi, B.; Finn, L.A.; Hagen, E.W.; Young, T.; Hla, K.M.; Van Cauter, E.; Peppard, P.E. Obstructive Sleep Apnea during REM Sleep and Hypertension. Results of the Wisconsin Sleep Cohort. Am. J. Respir. Crit. Care Med. 2014, 190, 1158–1167. [Google Scholar] [CrossRef] [PubMed]
- Newbold, R.; Benedetti, A.; Kimoff, R.J.; Meltzer, S.; Garfield, N.; Dasgupta, K.; Gagnon, R.; Lavigne, L.; Olha, A.; Rey, E.; et al. Maternal Sleep-Disordered Breathing in Pregnancy and Increased Nocturnal Glucose Levels in Women with Gestational Diabetes Mellitus. Chest 2021, 159, 356–365. [Google Scholar] [CrossRef]
- Balserak, B.I.; Pien, G.W.; Prasad, B.; Mastrogiannis, D.; Park, C.; Quinn, L.T.; Herdegen, J.; Carley, D.W. Obstructive Sleep Apnea Is Associated with Newly Diagnosed Gestational Diabetes Mellitus. Ann. Am. Thorac. Soc. 2020, 17, 754–761. [Google Scholar] [CrossRef]
- Hofman, P.L.; Cutfield, W.S.; Robinson, E.M.; Bergman, R.N.; Menon, R.K.; Sperling, M.A.; Gluckman, P.D. Insulin Resistance in Short Children with Intrauterine Growth Retardation. J. Clin. Endocrinol. Metab. 1997, 82, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Leon, D.A.; Lithell, H.O.; Vågerö, D.; Koupilová, I.; Mohsen, R.; Berglund, L.; Lithell, U.-B.; McKeigue, P.M. Reduced Fetal Growth Rate and Increased Risk of Death from Ischaemic Heart Disease: Cohort Study of 15,000 Swedish Men and Women Born 1915-29. BMJ 1998, 317, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Hincker, A.; Nadler, J.; Karan, S.; Carter, E.; Porat, S.; Warner, B.; Ju, Y.-E.S.; Ben Abdallah, A.; Wilson, E.; Lockhart, E.M.; et al. Sleep Apnea and Fetal Growth Restriction (SAFER) Study: Protocol for a Pragmatic Randomised Clinical Trial of Positive Airway Pressure as an Antenatal Therapy for Fetal Growth Restriction in Maternal Obstructive Sleep Apnoea. BMJ Open 2021, 11, e049120. [Google Scholar] [CrossRef] [PubMed]
- Kapral, N.; Miller, S.E.; Scharf, R.J.; Gurka, M.J.; DeBoer, M.D. Associations between Birthweight and Overweight and Obesity in School-Age Children. Pediatr. Obes. 2018, 13, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cai, R.; Lv, G.; Huang, Z.; Wang, Z. Hypoxia during Pregnancy in Rats Leads to the Changes of the Cerebral White Matter in Adult Offspring. Biochem. Biophys. Res. Commun. 2010, 396, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Brener, A.; Lebenthal, Y.; Levy, S.; Dunietz, G.L.; Sever, O.; Tauman, R. Mild Maternal Sleep-Disordered Breathing during Pregnancy and Offspring Growth and Adiposity in the First 3 Years of Life. Sci. Rep. 2020, 10, 13979. [Google Scholar] [CrossRef] [PubMed]
- Tauman, R.; Zuk, L.; Uliel-Sibony, S.; Ascher-Landsberg, J.; Katsav, S.; Farber, M.; Sivan, Y.; Bassan, H. The Effect of Maternal Sleep-Disordered Breathing on the Infant’s Neurodevelopment. Am. J. Obstet. Gynecol. 2015, 212, 656.e1–656.e7. [Google Scholar] [CrossRef] [PubMed]
- Morrakotkhiew, W.; Chirdkiatgumchai, V.; Tantrakul, V.; Thampratankul, L. Early Developmental Outcome in Children Born to Mothers with Obstructive Sleep Apnea. Sleep Med. 2021, 88, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Christensson, E.; Mkrtchian, S.; Ebberyd, A.; Österlund Modalen, Å.; Franklin, K.A.; Eriksson, L.I.; Jonsson Fagerlund, M. Whole Blood Gene Expression Signature in Patients with Obstructive Sleep Apnea and Effect of Continuous Positive Airway Pressure Treatment. Respir. Physiol. Neurobiol. 2021, 294, 103746. [Google Scholar] [CrossRef]
- Moreira, S.; Rodrigues, R.; Barros, A.B.; Pejanovic, N.; Neves-Costa, A.; Pedroso, D.; Pereira, C.; Fernandes, D.; Rodrigues, J.V.; Barbara, C.; et al. Changes in Expression of the CLOCK Gene in Obstructive Sleep Apnea Syndrome Patients Are Not Reverted by Continuous Positive Airway Pressure Treatment. Front. Med. 2017, 4, 285306. [Google Scholar] [CrossRef]
- Xie, T.; Guo, D.; Luo, J.; Guo, Z.; Zhang, S.; Wang, A.; Wang, X.; Wang, X.; Cao, W.; Su, L.; et al. The Relationship Between HIF1α and Clock Gene Expression in Patients with Obstructive Sleep Apnea. Nat. Sci. Sleep 2022, 14, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lu, Y.; Sheng, L.; Han, X.; Yu, L.; Zhang, W.; Liu, S.; Liu, Y. Advances in Molecular Pathology of Obstructive Sleep Apnea. Molecules 2022, 27, 8422. [Google Scholar] [CrossRef] [PubMed]
- Kidron, D.; Bar-Lev, Y.; Tsarfaty, I.; Many, A.; Tauman, R. The Effect of Maternal Obstructive Sleep Apnea on the Placenta. Sleep 2019, 42, zsz072. [Google Scholar] [CrossRef] [PubMed]
- Johns, E.C.; Halligan, D.L.; Tammsalu, T.; Hill, E.A.; Riha, R.L.; Denison, F.C.; Reynolds, R.M. Gene Expression Profiling of Placentae from Women with Obesity and Obstructive Sleep Apnoea. Placenta 2022, 121, 53–60. [Google Scholar] [CrossRef] [PubMed]
- de Knegt, V.E.; Hedley, P.L.; Kanters, J.K.; Thagaard, I.N.; Krebs, L.; Christiansen, M.; Lausten-Thomsen, U. The Role of Leptin in Fetal Growth during Pre-Eclampsia. Int. J. Mol. Sci. 2021, 22, 4569. [Google Scholar] [CrossRef] [PubMed]
- Valverde-Pérez, E.; Prieto-Lloret, J.; Gonzalez-Obeso, E.; Cabero, M.I.; Nieto, M.L.; Pablos, M.I.; Obeso, A.; Gomez-Niño, A.; Cárdaba-García, R.M.; Rocher, A.; et al. Effects of Gestational Intermittent Hypoxia on Placental Morphology and Fetal Development in a Murine Model of Sleep Apnea. Adv. Exp. Med. Biol. 2023, 1427, 73–81. [Google Scholar] [CrossRef]
- Chen, L.; Zadi, Z.H.; Zhang, J.; Scharf, S.M.; Pae, E.K. Intermittent Hypoxia in Utero Damages Postnatal Growth and Cardiovascular Function in Rats. J. Appl. Physiol. 2018, 124, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Weng, C.; Huang, L.; Feng, H.; He, Q.; Lin, X.; Jiang, T.; Lin, J.; Wang, X.; Liu, Q. Gestational Chronic Intermittent Hypoxia Induces Hypertension, Proteinuria, and Fetal Growth Restriction in Mice. Sleep Breath. 2022, 26, 1661–1669. [Google Scholar] [CrossRef]
- Sukarieh, R.; Joseph, R.; Leow, S.C.; Li, Y.; Löffler, M.; Aris, I.M.; Tan, J.H.; Teh, A.L.; Chen, L.; Holbrook, J.D.; et al. Molecular Pathways Reflecting Poor Intrauterine Growth Are Found in Wharton’s Jelly-Derived Mesenchymal Stem Cells. Hum. Reprod. 2014, 29, 2287–2301. [Google Scholar] [CrossRef]
- Locke, B.W.; Lee, J.J.; Sundar, K.M. OSA and Chronic Respiratory Disease: Mechanisms and Epidemiology. Int. J. Environ. Res. Public Health 2022, 19, 5473. [Google Scholar] [CrossRef]
- Meyer, E.J.; Wittert, G.A. Approach the Patient With Obstructive Sleep Apnea and Obesity. J. Clin. Endocrinol. Metab. 2024, 109, e1267–e1279. [Google Scholar] [CrossRef]
- Shobatake, R.; Ota, H.; Takahashi, N.; Ueno, S.; Sugie, K.; Takasawa, S. The Impact of Intermittent Hypoxia on Metabolism and Cognition. Int. J. Mol. Sci. 2022, 23, 12957. [Google Scholar] [CrossRef]
- Kapsimalis, F.; Kryger, M. Sleep Breathing Disorders in the U.S. Female Population. J. Women’s Health 2009, 18, 1211–1219. [Google Scholar] [CrossRef]
- Bonsignore, M.R.; Mazzuca, E.; Baiamonte, P.; Bouckaert, B.; Verbeke, W.; Pevernagie, D.A. REM Sleep Obstructive Sleep Apnoea. Eur. Respir. Rev. 2024, 33, 230166. [Google Scholar] [CrossRef]
- Mabry, S.; Wilson, E.N.; Bradshaw, J.L.; Gardner, J.J.; Fadeyibi, O.; Vera, E.; Osikoya, O.; Cushen, S.C.; Karamichos, D.; Goulopoulou, S.; et al. Sex and Age Differences in Social and Cognitive Function in Offspring Exposed to Late Gestational Hypoxia. Biol. Sex Differ. 2023, 14, 81. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, B.; Huang, Z.; Duan, R.; Li, H.; Xie, L.; Wang, R.; Li, Z.; Gao, Y.; Zheng, Y.; et al. Effects of Poor Sleep on the Immune Cell Landscape as Assessed by Single-Cell Analysis. Commun. Biol. 2021, 4, 1325. [Google Scholar] [CrossRef]
- Gluckman, P.D.; Hanson, M.A.; Cooper, C.; Thornburg, K.L. Effect of in Utero and Early-Life Conditions on Adult Health and Disease. N. Engl. J. Med. 2008, 359, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Liew, S.C.; Aung, T. Sleep Deprivation and Its Association with Diseases—A Review. Sleep Med. 2021, 77, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Alhendi, A.S.N.; Lim, D.; McKee, S.; McEntagart, M.; Tatton-Brown, K.; Temple, I.K.; Davies, J.H.; MacKay, D.J.G. Whole-Genome Analysis as a Diagnostic Tool for Patients Referred for Diagnosis of Silver-Russell Syndrome: A Real-World Study. J. Med. Genet. 2022, 59, 613–622. [Google Scholar] [CrossRef]
- Pagel, J.-I.; Deindl, E. Disease Progression Mediated by Egr-1 Associated Signaling in Response to Oxidative Stress. Int. J. Mol. Sci. 2012, 13, 13104–13117. [Google Scholar] [CrossRef]
- Louis, J.; Auckley, D.; Miladinovic, B.; Shepherd, A.; Mencin, P.; Kumar, D.; Mercer, B.; Redline, S. Perinatal Outcomes Associated with Obstructive Sleep Apnea in Obese Pregnant Women. Obstet. Gynecol. 2012, 120, 1085–1092. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology Knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Sato, Y.; Kawashima, M.; Kanehisa, M. KEGG Tools for Classification and Analysis of Viral Proteins. Protein Sci. 2023, 32, e4820. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Balcı, H.; Hanspers, K.; Coort, S.L.; Martens, M.; Slenter, D.N.; Ehrhart, F.; Digles, D.; Waagmeester, A.; Wassink, I.; et al. WikiPathways 2024: Next Generation Pathway Database. Nucleic Acids Res. 2024, 52, D679–D689. [Google Scholar] [CrossRef]
- Agapito, G.; Arbitrio, M. Microarray Data Analysis Protocol. Methods Mol. Biol. 2022, 2401, 263–271. [Google Scholar] [CrossRef]





| Microarray Cohort | qRT-PCR Cohort | |||||
|---|---|---|---|---|---|---|
| OSA (n = 5) | Non-OSA (n = 5) | p | OSA (n = 7) | Non OSA (n = 13) | p | |
| Age (years) | 36 (36–40) | 36 (33–37) | 0.42 | 36 (34–38) | 34 (33–37) | 0.64 |
| BMI before pregnancy (kg/m2) | 22.3 (21–24) | 21.9 (20.1–21.9) | 0.73 | 22.3 (20.8–24.8) | 21.7 (20.8–22.7) | 0.64 |
| Neck circumference (cm) | 34 (32–34.5) | 32 (34–37) | 0.53 | 34 (31.8–34.8) | 33.5 (32–34) | 0.94 |
| Waist (cm) | 95.5 (95–106) | 107 (96–112) | 0.35 | 99.5 (95.3–106.3) | 104 (96–108) | 0.49 |
| Hip circumference (cm) | 102 (99.25–104) | 107.5 (99–116) | 0.56 | 102 (99.3–104) | 106.5 (97–115) | 0.86 |
| Gestational age at PSG (weeks) | 36 (32.5–38) | 31 (30–33) | 0.11 | 34 (31–37) | 34 (31–35) | 0.77 |
| First pregnancy (n) | 3 | 1 | 0.52 | 4 | 5 | 0.64 |
| Gestational smokers (n) | 1 | 0 | 1 | 1 | 0 | 0.35 |
| Epworth Sleepiness Scale | 7 (6–9) | 4 (3–5) | 0.1 | 7 (5–8) | 6 (4–10) | 1 |
| AHI (h−1) | 6.4 (5.3–7.6) | 0.1 (0.0–0.2) | 0.01 | 6.4 (5.5–8.6) | 0.4 (0.1–0.7) | 0.00 |
| Desaturation index (h−1) | 0.3 (0–1.1) | 0 (0–0.1) | 0.05 | 0.6 (0.3–1.7) | 0 (0–0.4) | 0.01 |
| REM-AHI (h−1) | 10 (4.8–10.3) | 0 (0–1) | 0.05 | 10 (3.3–16.5) | 0 (0–1) | 0.00 |
| Birthweight (g) | 3180 (3015–3270) | 3210 (3200–3225) | 0.84 | 3180 (2967–3265) | 3232 (3080–3430) | 0.40 |
| Cesarean section (n) | 1 | 1 | 1 | 1 | 1 | 1 |
| Neonatal gestational age (weeks) | 39 (38–40) | 39 (38–40) | 1 | 39 (39–40) | 40 (38–40) | 0.39 |
| Selection of Primers | Reference Sequence ID | Exon Boundary | Forward | Reverse |
|---|---|---|---|---|
| 18S rRNA | NR_145820.1 | 5′-TAAGCAACGAGACTCTGGCAT | 5′-CGGACATCTAAGGGCATCACAG | |
| AKT2 | NM_001243027 | 2–3 | 5′-ACTTCCTGCTGAAGAGCGAC | 5′-CCTCCCTCTCGTCTGGAGAA |
| CAT | NM_001752 | 12–13 | 5′-CTCTTCTGGACAAGTACAATG | 5′-AGGAGAATCTTCATCCAGTG |
| EGR1 | NM_001964 | 1–2 | 5′-GCAGAGTCTTTTCCTGAC | 5′-TTGGTCATGCTCACTAGG |
| IGF2 | NM_000612 | 3–4 | 5′-GGACAACTTCCCCAGATAC | 5′-GTGGGTAGAGCAATCAGG |
| JUNB | NM_002229 | NA | 5′-TACTGTGGAAAAGAAACACG | 5′-GAACAAACACACACAAACAC |
| MAPKAP1 | NM_001006617 | 6–7 | 5′-TAAATGCTGCTCATGGATTC | 5′-AACTTTCTGGGATCCTTTTC |
| MCL1 | NM_001197320 | 3–4 | 5′-TAGTTAAACAAAGAGGCTGG | 5′-ATAAACTGGTTTTGGTGGTG |
| MYC | NM_002467 | 2–3 | 5′-TGAGGAGGAACAAGAAGATG | 5′-ATCCAGACTCTGACCTTTTG |
| PFN1 | NM_005022 | 2–3 | 5′-TTCTTGTTGATCAAACCACC | 5′-GGGAATTTAGCATGGATCTTC |
| PPP1CB | NM_002709 | NA | 5′-GACCATAGCAAATCACAGAG | 5′-GCAATCTATGGAGCAGATTC |
| PRKAR1A | NM_002734 | 6–7 | 5′-GAAGGGGATAACTTCTATGT | 5′-ATTCTTCTATAGCTGTCTCGG |
| STX4 | NM_004604 | 9–10 | 5′-GATTGAGAAGAACATCCTGAG | 5′-CATTATCCAACCACTGTGAC |
| UBA52 | NM_001033930 | 4–5 | 5′-CCTTATTTGACCTTCTTCTTGG | 5′-CTTGCCCAGAAATACAACTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cànaves-Gómez, L.; Fleischer, A.; Muncunill-Farreny, J.; Gimenez, M.P.; Álvarez Ruiz De Larrinaga, A.; Sánchez Baron, A.; Codina Marcet, M.; De-La-Peña, M.; Morell-Garcia, D.; Peña Zarza, J.; et al. Effect of Obstructive Sleep Apnea during Pregnancy on Fetal Development: Gene Expression Profile of Cord Blood. Int. J. Mol. Sci. 2024, 25, 5537. https://doi.org/10.3390/ijms25105537
Cànaves-Gómez L, Fleischer A, Muncunill-Farreny J, Gimenez MP, Álvarez Ruiz De Larrinaga A, Sánchez Baron A, Codina Marcet M, De-La-Peña M, Morell-Garcia D, Peña Zarza J, et al. Effect of Obstructive Sleep Apnea during Pregnancy on Fetal Development: Gene Expression Profile of Cord Blood. International Journal of Molecular Sciences. 2024; 25(10):5537. https://doi.org/10.3390/ijms25105537
Chicago/Turabian StyleCànaves-Gómez, Laura, Aarne Fleischer, Josep Muncunill-Farreny, María Paloma Gimenez, Ainhoa Álvarez Ruiz De Larrinaga, Andrés Sánchez Baron, Mercedes Codina Marcet, Mónica De-La-Peña, Daniel Morell-Garcia, José Peña Zarza, and et al. 2024. "Effect of Obstructive Sleep Apnea during Pregnancy on Fetal Development: Gene Expression Profile of Cord Blood" International Journal of Molecular Sciences 25, no. 10: 5537. https://doi.org/10.3390/ijms25105537




