Cross-Talks between Raf Kinase Inhibitor Protein and Programmed Cell Death Ligand 1 Expressions in Cancer: Role in Immune Evasion and Therapeutic Implications
Abstract
:1. Introduction
1.1. The PD-L1/PD-1 Pathway in Immune Evasion
1.2. PD-L1 and Inactivation of CD8+ T Cells
1.3. Clinical Implications of the PD-1/PD-L1 Axis
2. RKIP Properties and Immune Activation
2.1. RKIP Signaling Pathways
2.2. Cross-Talks between RKIP and PD-L1
2.3. Cross-Talk via the MAPK Pathway
2.4. Cross-Talk via Cytokines IL-1β and IFN-γ
2.5. Cross-Talk via GSK3β
2.6. Cross-Talk via the Sox2 Oncogene
2.7. Cross-Talk via YY1 and NFκB
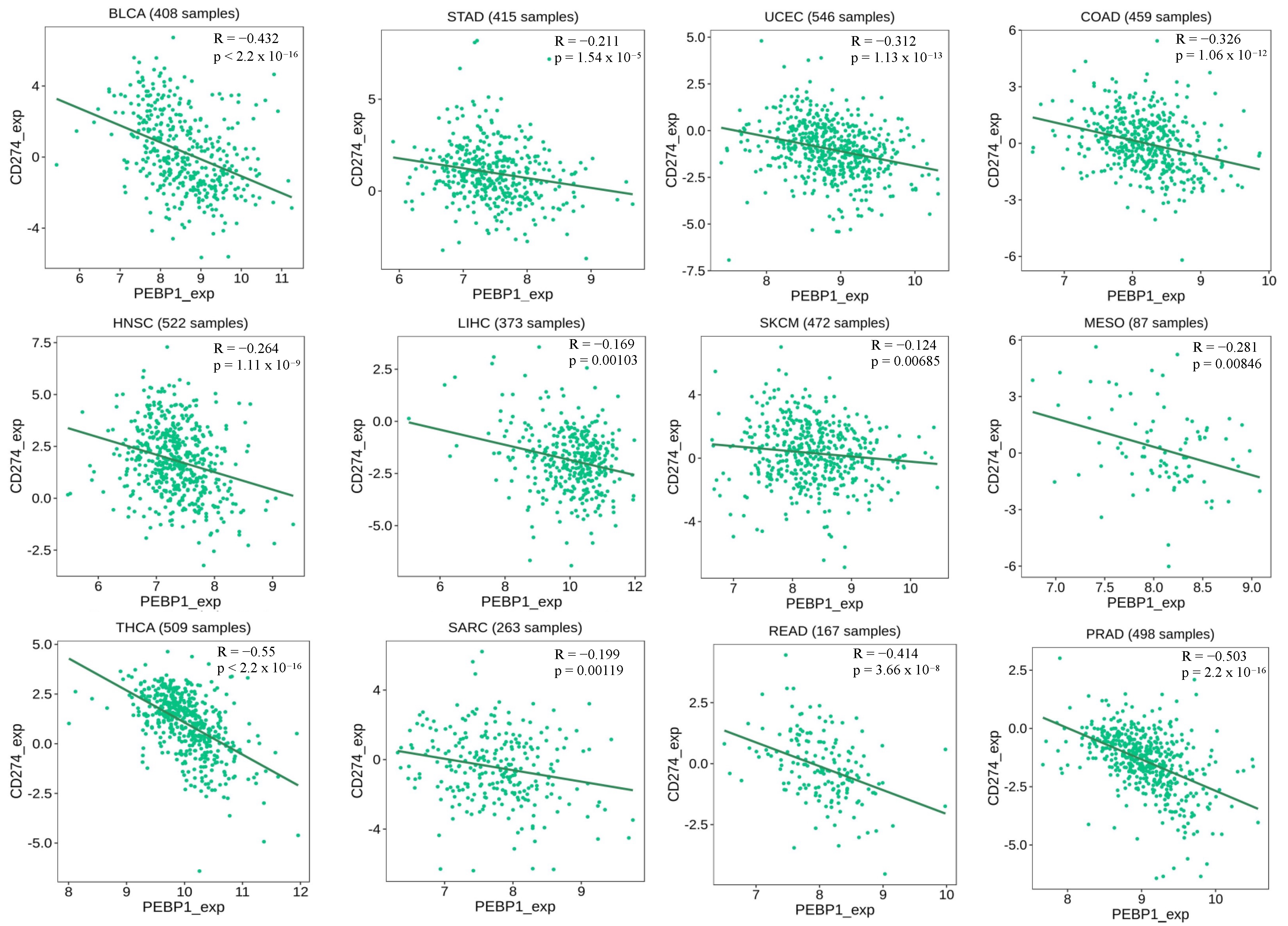
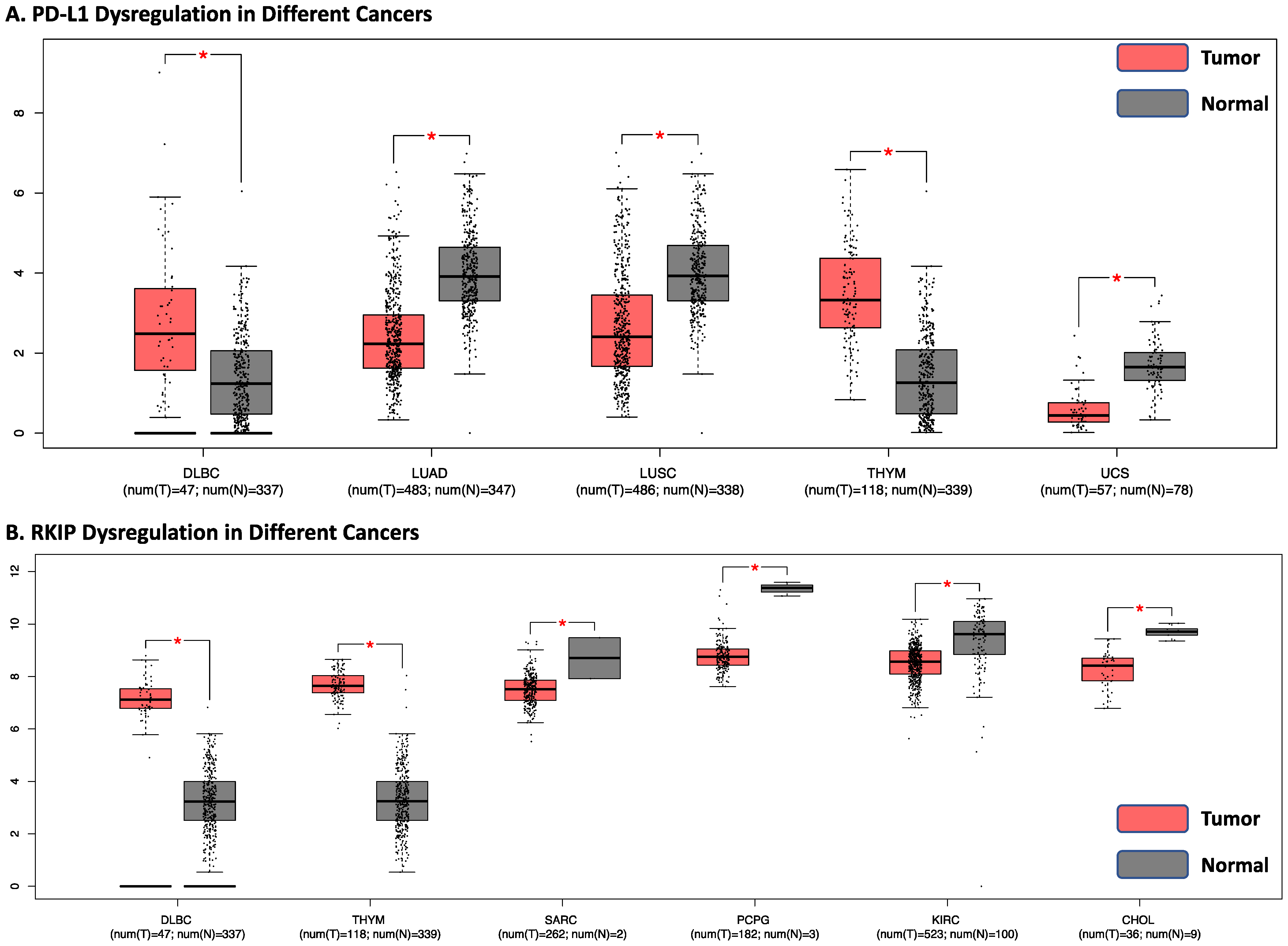
2.8. Analyses of the Correlation between RKIP and PD-L1 Expressions in Human Cancers by Bioinformatics
2.9. Potential Therapeutic Strategies Targeting RKIP and PD-L1
RKIP Inducers
2.10. PD-L1 Inhibitors
2.11. Combining Targeting Both RKIP and PD-L1
3. Clinical Trials
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| RKIP | Raf Kinase Inhibitor Protein |
| ICIs | Immune checkpoint inhibitors |
| TME | Tumor microenvironment |
| PD-1 | Programmed cell death 1 receptor |
| PD-L1 and PD-L2 | Programmed cell death ligand 1 and 2 |
| PEBP | Phosphatidylethanolamine-binding protein |
| MAPK | Mitogen-activated protein kinase |
| SOX2 | Sex determining region Y-box 2 |
| ITIM | Immunoreceptor tyrosine-based inhibitory motif |
| ITSM | Immunoreceptor tyrosine-based switch motif |
| SHP1 and SHP2 | Src homology domain 1 and/or 2 containing phosphatases |
| NF-κB | Nuclear factor kappa B |
| PI3K | Phosphoinositide 3-kinase/protein kinase |
| PTEN | Phosphatase and tensin homolog |
| IFNγ | Interferon-γ |
| pRKIP | Phosphorylated RKIP |
| TAK-1 | Growth factor β-activated kinase 1 |
| GSK3β | Glycogen synthase kinase-3β |
| mAbs | Monoclonal antibodies |
| TGFβ | Transforming growth factor-beta |
| EGCG | Epigallocatechin gallate |
| JAK1 and JAK2 | Janus kinases 1 and 2 |
| STAT3 | Signal transducer and activator of transcription 3 |
| IL-1β | Interleukin-1 beta |
References
- Abbott, M.; Ustoyev, Y. Cancer and the Immune System: The History and Background of Immunotherapy. Semin. Oncol. Nurs. 2019, 35, 150923. [Google Scholar] [CrossRef] [PubMed]
- Olszanski, A.J. Principles of Immunotherapy. J. Natl. Compr. Cancer Netw. 2015, 13, 670–672. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.-R.; Corrales, L.; Gajewski, T.F. Innate Immune Recognition of Cancer. Annu. Rev. Immunol. 2015, 33, 445–474. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Jeon, J.W.; Sievers, C.; Allen, C.T. Antigen Processing and Presentation in Cancer Immunotherapy. J. Immunother. Cancer 2020, 8, e001111. [Google Scholar] [CrossRef] [PubMed]
- Vesely, M.D.; Kershaw, M.H.; Schreiber, R.; Smyth, M. Natural Innate and Adaptive Immunity to Cancer. Annu. Rev. Immunol. 2011, 29, 235–271. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Song, Y.; Du, W.; Gong, L.; Chang, H.; Zou, Z. Tumor-Associated Macrophages: An Accomplice in Solid Tumor Progression. J. Biomed. Sci. 2019, 26, 78. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, Z.; Ding, Y.; Qin, Y. Tumor Microenvironment-Mediated Immune Evasion in Hepatocellular Carcinoma. Front. Immunol. 2023, 14, 1133308. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.M.; Marabelle, A.; Eggermont, A.; Soria, J.-C.; Kroemer, G.; Zitvogel, L. Targeting the Tumor Microenvironment: Removing Obstruction to Anticancer Immune Responses and Immunotherapy. Ann. Oncol. 2016, 27, 1482–1492. [Google Scholar] [CrossRef]
- Jin, H.-T.; Anderson, A.C.; Tan, W.G.; West, E.E.; Ha, S.-J.; Araki, K.; Freeman, G.J.; Kuchroo, V.K.; Ahmed, R. Cooperation of Tim-3 and PD-1 in CD8 T-Cell Exhaustion during Chronic Viral Infection. Proc. Natl. Acad. Sci. USA 2010, 107, 14733–14738. [Google Scholar] [CrossRef]
- Raskov, H.; Orhan, A.; Christensen, J.P.; Gögenur, I. Cytotoxic CD8+ T Cells in Cancer and Cancer Immunotherapy. Br. J. Cancer 2021, 124, 359–367. [Google Scholar] [CrossRef]
- Yin, Z.; Zhang, Y.; Wang, X. Advances in Chimeric Antigen Receptor T-Cell Therapy for B-Cell Non-Hodgkin Lymphoma. Biomark. Res. 2021, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Maalej, K.M.; Merhi, M.; Inchakalody, V.P.; Mestiri, S.; Alam, M.; Maccalli, C.; Cherif, H.; Uddin, S.; Steinhoff, M.; Marincola, F.M.; et al. CAR-Cell Therapy in the Era of Solid Tumor Treatment: Current Challenges and Emerging Therapeutic Advances. Mol. Cancer 2023, 22, 20. [Google Scholar] [CrossRef]
- Granhøj, J.S.; Witness Præst Jensen, A.; Presti, M.; Met, Ö.; Svane, I.M.; Donia, M. Tumor-Infiltrating Lymphocytes for Adoptive Cell Therapy: Recent Advances, Challenges, and Future Directions. Expert Opin. Biol. Ther. 2022, 22, 627–641. [Google Scholar] [CrossRef]
- Met, Ö.; Jensen, K.M.; Chamberlain, C.A.; Donia, M.; Svane, I.M. Principles of Adoptive T Cell Therapy in Cancer. Semin. Immunopathol. 2019, 41, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Wolchok, J.D.; Chen, L. PD-L1 (B7-H1) and PD-1 Pathway Blockade for Cancer Therapy: Mechanisms, Response Biomarkers, and Combinations. Sci. Transl. Med. 2016, 8, 328rv4. [Google Scholar] [CrossRef]
- Rowshanravan, B.; Halliday, N.; Sansom, D.M. CTLA-4: A Moving Target in Immunotherapy. Blood 2018, 131, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Ge, J.; Xiang, B.; Wu, X.; Ma, J.; Zhou, M.; Li, X.; et al. Role of the Tumor Microenvironment in PD-L1/PD-1-Mediated Tumor Immune Escape. Mol. Cancer 2019, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, T.; Honjo, T. PD-1 and PD-1 Ligands: From Discovery to Clinical Application. Int. Immunol. 2007, 19, 813–824. [Google Scholar] [CrossRef]
- Ribas, A.; Wolchok, J.D. Cancer Immunotherapy Using Checkpoint Blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef]
- Zhang, K.; Kong, X.; Li, Y.; Wang, Z.; Zhang, L.; Xuan, L. PD-1/PD-L1 Inhibitors in Patients with Preexisting Autoimmune Diseases. Front. Pharmacol. 2022, 13, 854967. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.P.; Kurzrock, R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol. Cancer Ther. 2015, 14, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Imamoto, A.; Rosner, M.R. Raf Kinase Inhibitory Protein (RKIP): A Physiological Regulator and Future Therapeutic Target. Expert Opin. Ther. Targets 2008, 12, 1275–1287. [Google Scholar] [CrossRef] [PubMed]
- Klysik, J.; Theroux, S.J.; Sedivy, J.M.; Moffit, J.S.; Boekelheide, K. Signaling Crossroads: The Function of Raf Kinase Inhibitory Protein in Cancer, the Central Nervous System and Reproduction. Cell. Signal. 2008, 20, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yeung, K.; Seitz, T.; Li, S.; Janosch, P.; McFerran, B.; Kaiser, C.; Fee, F.; Katsanakis, K.D.; Rose, D.W.; Mischak, H.; et al. Suppression of Raf-1 Kinase Activity and MAP Kinase Signalling by RKIP. Nature 1999, 401, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Xu, H.; Zhou, Y.; Wu, Z.; Jiang, B.; Chen, H.; Lin, D. Cefotetan-Bound Human RKIP Involves in Ras/Raf1/MEK/ERK Signaling Pathway. Acta Biochim. Biophys. Sin. 2022, 54, 1917–1923. [Google Scholar] [CrossRef] [PubMed]
- Papale, M.; Netti, G.S.; Stallone, G.; Ranieri, E. Understanding Mechanisms of RKIP Regulation to Improve the Development of New Diagnostic Tools. Cancers 2022, 14, 5070. [Google Scholar] [CrossRef] [PubMed]
- Gabriela-Freitas, P.; Pinheiro, J.; Raquel-Cunha, A.; Cardoso-Carneiro, D. Martinho, O. RKIP as an Inflammatory and Immune System Modulator: Implications in Cancer. Biomolecules 2019, 9, 769. [Google Scholar] [CrossRef]
- Lamiman, K.; Keller, J.M.; Mizokami, A.; Zhang, J.; Keller, E.T. Survey of Raf Kinase Inhibitor Protein (RKIP) in Multiple Cancer Types. Crit. Rev. Oncog. 2014, 19, 455–468. [Google Scholar] [CrossRef]
- Wang, X.; Yang, L.; Huang, F.; Zhang, Q.; Liu, S.; Ma, L.; You, Z. Inflammatory Cytokines IL-17 and TNF-α up-Regulate PD-L1 Expression in Human Prostate and Colon Cancer Cells. Immunol. Lett. 2017, 184, 7–14. [Google Scholar] [CrossRef]
- Boussiotis, V.A.; Chatterjee, P.; Li, L. Biochemical Signaling of PD-1 on T Cells and Its Functional Implications. Cancer J. 2014, 20, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Patsoukis, N.; Duke-Cohan, J.S.; Chaudhri, A.; Aksoylar, H.-I.; Wang, Q.; Council, A.; Berg, A.; Freeman, G.J.; Boussiotis, V.A. Interaction of SHP-2 SH2 Domains with PD-1 ITSM Induces PD-1 Dimerization and SHP-2 Activation. Commun. Biol. 2020, 3, 128. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Masubuchi, T.; Cai, Q.; Zhao, Y.; Hui, E. Molecular Features Underlying Differential SHP1/SHP2 Binding of Immune Checkpoint Receptors. eLife 2021, 10, e74276. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, T.; Maeda, A.; Nishimura, H.; Kurosaki, T.; Honjo, T. PD-1 Immunoreceptor Inhibits B Cell Receptor-Mediated Signaling by Recruiting Src Homology 2-Domain-Containing Tyrosine Phosphatase 2 to Phosphotyrosine. Proc. Natl. Acad. Sci. USA 2001, 98, 13866–13871. [Google Scholar] [CrossRef] [PubMed]
- Boussiotis, V.A.; Patsoukis, N. Effects of PD-1 Signaling on Immunometabolic Reprogramming. Immunometabolism 2022, 4, 220007. [Google Scholar] [CrossRef] [PubMed]
- Hofmeyer, K.A.; Jeon, H.; Zang, X. The PD-1/PD-L1 (B7-H1) Pathway in Chronic Infection-Induced Cytotoxic T Lymphocyte Exhaustion. J. Biomed. Biotechnol. 2011, 2011, 451694. [Google Scholar] [CrossRef] [PubMed]
- Siska, P.J.; Rathmell, J.C. T Cell Metabolic Fitness in Antitumor Immunity. Trends Immunol. 2015, 36, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Yokosuka, T.; Takamatsu, M.; Kobayashi-Imanishi, W.; Hashimoto-Tane, A.; Azuma, M.; Saito, T. Programmed Cell Death 1 Forms Negative Costimulatory Microclusters That Directly Inhibit T Cell Receptor Signaling by Recruiting Phosphatase SHP2. J. Exp. Med. 2012, 209, 1201–1217. [Google Scholar] [CrossRef]
- Papa, A.; Pandolfi, P.P. The PTEN–PI3K Axis in Cancer. Biomolecules 2019, 9, 153. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Gu, Z.; Chen, Y.; Chen, B.; Chen, W.; Weng, L.; Liu, X. Application of PD-1 Blockade in Cancer Immunotherapy. Comput. Struct. Biotechnol. J. 2019, 17, 661–674. [Google Scholar] [CrossRef]
- Tang, H.; Liang, Y.; Anders, R.A.; Taube, J.M.; Qiu, X.; Mulgaonkar, A.; Liu, X.; Harrington, S.M.; Guo, J.; Xin, Y.; et al. PD-L1 on Host Cells Is Essential for PD-L1 Blockade–Mediated Tumor Regression. J. Clin. Investig. 2018, 128, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.-H.; Chan, L.-C.; Li, C.-W.; Hsu, J.L.; Hung, M.-C. Mechanisms Controlling PD-L1 Expression in Cancer. Mol. Cell 2019, 76, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Ivan, C.; Valdecanas, D.; Wang, X.; Peltier, H.J.; Ye, Y.; Araujo, L.; Carbone, D.P.; Shilo, K.; Giri, D.K.; et al. PDL1 Regulation by P53 via miR-34. JNCI J. Natl. Cancer Inst. 2016, 108, djv303. [Google Scholar] [CrossRef]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on Tumor Cells in the Escape from Host Immune System and Tumor Immunotherapy by PD-L1 Blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [PubMed]
- McLane, L.M.; Abdel-Hakeem, M.S.; Wherry, E.J. CD8 T Cell Exhaustion during Chronic Viral Infection and Cancer. Annu. Rev. Immunol. 2019, 37, 457–495. [Google Scholar] [CrossRef]
- Dolina, J.S.; Van Braeckel-Budimir, N.; Thomas, G.D.; Salek-Ardakani, S. CD8+ T Cell Exhaustion in Cancer. Front. Immunol. 2021, 12, 715234. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liang, X.; Dong, W.; Fang, Y.; Lv, J.; Zhang, T.; Fiskesund, R.; Xie, J.; Liu, J.; Yin, X.; et al. Tumor-Repopulating Cells Induce PD-1 Expression in CD8+ T Cells by Transferring Kynurenine and AhR Activation. Cancer Cell 2018, 33, 480–494.e7. [Google Scholar] [CrossRef] [PubMed]
- Ahn, E.; Araki, K.; Hashimoto, M.; Li, W.; Riley, J.L.; Cheung, J.; Sharpe, A.H.; Freeman, G.J.; Irving, B.A.; Ahmed, R. Role of PD-1 during Effector CD8 T Cell Differentiation. Proc. Natl. Acad. Sci. USA 2018, 115, 4749–4754. [Google Scholar] [CrossRef] [PubMed]
- Wherry, E.J.; Ha, S.-J.; Kaech, S.M.; Haining, W.N.; Sarkar, S.; Kalia, V.; Subramaniam, S.; Blattman, J.N.; Barber, D.L.; Ahmed, R. Molecular Signature of CD8+ T Cell Exhaustion during Chronic Viral Infection. Immunity 2007, 27, 670–684. [Google Scholar] [CrossRef]
- Gong, J.; Chehrazi-Raffle, A.; Reddi, S.; Salgia, R. Development of PD-1 and PD-L1 Inhibitors as a Form of Cancer Immunotherapy: A Comprehensive Review of Registration Trials and Future Considerations. J. Immunother. Cancer 2018, 6, 8. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, M.; Nie, H.; Yuan, Y. PD-1 and PD-L1 in Cancer Immunotherapy: Clinical Implications and Future Considerations. Hum. Vaccines Immunother. 2019, 15, 1111–1122. [Google Scholar] [CrossRef]
- Winer, A.; Ghatalia, P.; Bubes, N.; Anari, F.; Varshavsky, A.; Kasireddy, V.; Liu, Y.; El-Deiry, W.S. Dual Checkpoint Inhibition with Ipilimumab plus Nivolumab After Progression on Sequential PD-1/PDL-1 Inhibitors Pembrolizumab and Atezolizumab in a Patient with Lynch Syndrome, Metastatic Colon, and Localized Urothelial Cancer. Oncologist 2019, 24, 1416–1419. [Google Scholar] [CrossRef] [PubMed]
- Bernier, I.; Jollés, P. Purification and Characterization of a Basic 23 kDa Cytosolic Protein from Bovine Brain. Biochim. Biophys. Acta BBA-Protein Struct. Mol. Enzymol. 1984, 790, 174–181. [Google Scholar] [CrossRef]
- Bonavida, B. RKIP-Mediated Chemo-Immunosensitization of Resistant Cancer Cells via Disruption of the NF-κB/Snail/YY1/RKIP Resistance-Driver Loop. Crit. Rev. Oncog. 2014, 19, 431–445. [Google Scholar] [CrossRef]
- Datar, I.; Qiu, X.; Ma, H.Z.; Yeung, M.; Aras, S.; De La Serna, I.; Al-Mulla, F.; Thiery, J.P.; Trumbly, R.; Fan, X.; et al. RKIP Regulates CCL5 Expression to Inhibit Breast Cancer Invasion and Metastasis by Controlling Macrophage Infiltration. Oncotarget 2015, 6, 39050–39061. [Google Scholar] [CrossRef]
- Deiss, K.; Kisker, C.; Lohse, M.J.; Lorenz, K. Raf Kinase Inhibitor Protein (RKIP) Dimer Formation Controls Its Target Switch from Raf1 to G Protein-Coupled Receptor Kinase (GRK) 2. J. Biol. Chem. 2012, 287, 23407–23417. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, K.; Rosner, M.R. Harnessing RKIP to Combat Heart Disease and Cancer. Cancers 2022, 14, 867. [Google Scholar] [CrossRef] [PubMed]
- Cardile, V.; Malaponte, G.; Loreto, C.; Libra, M.; Caggia, S.; Trovato, F.M.; Musumeci, G. Raf Kinase Inhibitor Protein (RKIP) and Phospho-RKIP Expression in Melanomas. Acta Histochem. 2013, 115, 795–802. [Google Scholar] [CrossRef]
- Granovsky, A.E.; Clark, M.C.; McElheny, D.; Heil, G.; Hong, J.; Liu, X.; Kim, Y.; Joachimiak, G.; Joachimiak, A.; Koide, S.; et al. Raf Kinase Inhibitory Protein Function Is Regulated via a Flexible Pocket and Novel Phosphorylation-Dependent Mechanism. Mol. Cell. Biol. 2009, 29, 1306–1320. [Google Scholar] [CrossRef]
- Banfield, M.J.; Barker, J.J.; Perry, A.C.; Brady, R.L. Function from Structure? The Crystal Structure of Human Phosphatidylethanolamine-Binding Protein Suggests a Role in Membrane Signal Transduction. Structure 1998, 6, 1245–1254. [Google Scholar] [CrossRef]
- Skinner, J.J.; Rosner, M.R. RKIP Structure Drives Its Function: A Three-State Model for Regulation of RKIP. Crit. Rev. Oncog. 2014, 19, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, K.; Lohse, M.J.; Quitterer, U. Protein Kinase C Switches the Raf Kinase Inhibitor from Raf-1 to GRK-2. Nature 2003, 426, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Skinner, J.J.; Wang, S.; Lee, J.; Ong, C.; Sommese, R.; Sivaramakrishnan, S.; Koelmel, W.; Hirschbeck, M.; Schindelin, H.; Kisker, C.; et al. Conserved Salt-Bridge Competition Triggered by Phosphorylation Regulates the Protein Interactome. Proc. Natl. Acad. Sci. USA 2017, 114, 13453–13458. [Google Scholar] [CrossRef]
- Park, S.; Rath, O.; Beach, S.; Xiang, X.; Kelly, S.M.; Luo, Z.; Kolch, W.; Yeung, K.C. Regulation of RKIP Binding to the N-region of the Raf-1 Kinase. FEBS Lett. 2006, 580, 6405–6412. [Google Scholar] [CrossRef] [PubMed]
- Trakul, N.; Menard, R.E.; Schade, G.R.; Qian, Z.; Rosner, M.R. Raf Kinase Inhibitory Protein Regulates Raf-1 but Not B-Raf Kinase Activation. J. Biol. Chem. 2005, 280, 24931–24940. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yang, X.; Geng, M.; Huang, M. Targeting ERK, an Achilles’ Heel of the MAPK Pathway, in Cancer Therapy. Acta Pharm. Sin. B 2018, 8, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Yesilkanal, A.E.; Rosner, M.R. Raf Kinase Inhibitory Protein (RKIP) as a Metastasis Suppressor: Regulation of Signaling Networks in Cancer. Crit. Rev. Oncog. 2014, 19, 447–454. [Google Scholar] [CrossRef]
- Huerta-Yepez, S.; Yoon, N.K.; Hernandez-Cueto, A.; Mah, V.; Rivera-Pazos, C.M.; Chatterjee, D.; Vega, M.I.; Maresh, E.L.; Horvath, S.; Chia, D.; et al. Expression of Phosphorylated Raf Kinase Inhibitor Protein (pRKIP) Is a Predictor of Lung Cancer Survival. BMC Cancer 2011, 11, 259. [Google Scholar] [CrossRef]
- Li, S.; Liu, T.; Mo, W.; Hou, Q.; Zhou, Y.; Liu, M.; He, Z.; Liu, Z.; Chen, Q.; Wang, H.; et al. Prognostic Value of Phosphorylated Raf Kinase Inhibitory Protein at Serine 153 and Its Predictive Effect on the Clinical Response to Radiotherapy in Nasopharyngeal Carcinoma. Radiat. Oncol. 2016, 11, 121. [Google Scholar] [CrossRef]
- Zaravinos, A.; Bonavida, B.; Chatzaki, E.; Baritaki, S. RKIP: A Key Regulator in Tumor Metastasis Initiation and Resistance to Apoptosis: Therapeutic Targeting and Impact. Cancers 2018, 10, 287. [Google Scholar] [CrossRef]
- Cross-Knorr, S.; Lu, S.; Perez, K.; Guevara, S.; Brilliant, K.; Pisano, C.; Quesenberry, P.J.; Resnick, M.B.; Chatterjee, D. RKIP Phosphorylation and STAT3 Activation Is Inhibited by Oxaliplatin and Camptothecin and Are Associated with Poor Prognosis in Stage II Colon Cancer Patients. BMC Cancer 2013, 13, 463. [Google Scholar] [CrossRef] [PubMed]
- Shvartsur, A.; Givechian, K.B.; Garban, H.; Bonavida, B. Overexpression of RKIP and Its Cross-Talk with Several Regulatory Gene Products in Multiple Myeloma. J. Exp. Clin. Cancer Res. 2017, 36, 62. [Google Scholar] [CrossRef] [PubMed]
- Touboul, R.; Baritaki, S.; Zaravinos, A.; Bonavida, B. RKIP Pleiotropic Activities in Cancer and Inflammatory Diseases: Role in Immunity. Cancers 2021, 13, 6247. [Google Scholar] [CrossRef] [PubMed]
- Yeung, K.C.; Rose, D.W.; Dhillon, A.S.; Yaros, D.; Gustafsson, M.; Chatterjee, D.; McFerran, B.; Wyche, J.; Kolch, W.; Sedivy, J.M. Raf Kinase Inhibitor Protein Interacts with NF-κB-Inducing Kinase and TAK1 and Inhibits NF-κB Activation. Mol. Cell. Biol. 2001, 21, 7207–7217. [Google Scholar] [CrossRef]
- Bonavida, B.; Baritaki, S. Dual Role of NO Donors in the Reversal of Tumor Cell Resistance and EMT: Downregulation of the NF-κB/Snail/YY1/RKIP Circuitry. Nitric Oxide 2011, 24, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Escara-Wilke, J.; Yeung, K.; Keller, E.T. Raf Kinase Inhibitor Protein (RKIP) in Cancer. Cancer Metastasis Rev. 2012, 31, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Cessna, H.; Baritaki, S.; Zaravinos, A.; Bonavida, B. The Role of RKIP in the Regulation of EMT in the Tumor Microenvironment. Cancers 2022, 14, 4596. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Bonavida, B. The Activated NF-ĸB-Snail-RKIP Circuitry in Cancer Regulates Both the Metastatic Cascade and Resistance to Apoptosis by Cytotoxic Drugs. Crit. Rev. Oncog. 2009, 29, 241–254. [Google Scholar]
- Kujawski, M.; Kortylewski, M.; Lee, H.; Herrmann, A.; Kay, H.; Yu, H. Stat3 Mediates Myeloid Cell–Dependent Tumor Angiogenesis in Mice. J. Clin. Investig. 2008, 118, 3367–3377. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in Cancer Inflammation and Immunity: A Leading Role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Priceman, S.J.; Kujawski, M.; Shen, S.; Cherryholmes, G.A.; Lee, H.; Zhang, C.; Kruper, L.; Mortimer, J.; Jove, R.; Riggs, A.D.; et al. Regulation of Adipose Tissue T Cell Subsets by Stat3 Is Crucial for Diet-Induced Obesity and Insulin Resistance. Proc. Natl. Acad. Sci. USA 2013, 110, 13079–13084. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 Signalling in Cancer: New and Unexpected Biological Functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef]
- Vandamme, D.; Herrero, A.; Al-Mulla, F.; Kolch, W. Regulation of the MAPK Pathway by Raf Kinase Inhibitory Protein. Crit. Rev. Oncog. 2014, 19, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Wang, N.; Zhou, K.; Su, F.; Jiang, Y.; Shou, J.; Liu, H.; Ma, C.; Qian, Y.; Wang, K.; et al. RKIP Mediates Autoimmune Inflammation by Positively Regulating IL-17R Signaling. EMBO Rep. 2018, 19, e44951. [Google Scholar] [CrossRef]
- Bach, V.N.; Ding, J.; Yeung, M.; Conrad, T.; Odeh, H.N.; Cubberly, P.; Figy, C.; Ding, H.-F.; Trumbly, R.; Yeung, K.C. A Negative Regulatory Role for RKIP in Breast Cancer Immune Response. Cancers 2022, 14, 3605. [Google Scholar] [CrossRef]
- Christofi, T.; Zaravinos, A. RKIP in Human Diseases and Its Potential as a Prognostic Indicator and Therapeutic Target. In Prognostic and Therapeutic Applications of RKIP in Cancer; Elsevier: Amsterdam, The Netherlands, 2020; pp. 337–356. ISBN 978-0-12-819612-0. [Google Scholar]
- Wang, Y.; Bonavida, B. A New Linkage between the Tumor Suppressor RKIP and Autophagy: Targeted Therapeutics. Crit. Rev. Oncog. 2018, 23, 281–305. [Google Scholar] [CrossRef]
- Frankenberger, C.; Rabe, D.; Bainer, R.; Sankarasharma, D.; Chada, K.; Krausz, T.; Gilad, Y.; Becker, L.; Rosner, M.R. Metastasis Suppressors Regulate the Tumor Microenvironment by Blocking Recruitment of Prometastatic Tumor-Associated Macrophages. Cancer Res. 2015, 75, 4063–4073. [Google Scholar] [CrossRef]
- Zhao, J.; Wenzel, S. Interactions of RKIP with Inflammatory Signaling Pathways. Crit. Rev. Oncog. 2014, 19, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.; Alam, M.; Kashyap, T.; Nath, N.; Kumari, A.; Pramanik, K.K.; Nagini, S.; Mishra, R. Crosstalk between PD-L1 and Jak2-Stat3/MAPK-AP1 Signaling Promotes Oral Cancer Progression, Invasion and Therapy Resistance. Int. Immunopharmacol. 2023, 124, 110894. [Google Scholar] [CrossRef]
- Ribas, A.; Algazi, A.; Ascierto, P.A.; Butler, M.O.; Chandra, S.; Gordon, M.; Hernandez-Aya, L.; Lawrence, D.; Lutzky, J.; Miller, W.H.; et al. PD-L1 Blockade in Combination with Inhibition of MAPK Oncogenic Signaling in Patients with Advanced Melanoma. Nat. Commun. 2020, 11, 6262. [Google Scholar] [CrossRef]
- Stutvoet, T.S.; Kol, A.; De Vries, E.G.; De Bruyn, M.; Fehrmann, R.S.; Terwisscha Van Scheltinga, A.G.; De Jong, S. MAPK Pathway Activity Plays a Key Role in PD-L1 Expression of Lung Adenocarcinoma Cells. J. Pathol. 2019, 249, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Niu, M.; Xu, L.; Luo, S.; Wu, K. Regulation of PD-L1 Expression in the Tumor Microenvironment. J. Hematol. Oncol. 2021, 14, 10. [Google Scholar] [CrossRef]
- Hirayama, A.; Tanaka, K.; Tsutsumi, H.; Nakanishi, T.; Yamashita, S.; Mizusaki, S.; Ishii, Y.; Ota, K.; Yoneshima, Y.; Iwama, E.; et al. Regulation of PD-L1 Expression in Non–Small Cell Lung Cancer by Interleukin-1β. Front. Immunol. 2023, 14, 1192861. [Google Scholar] [CrossRef]
- Liu, L.; Sun, Q.; Bao, R.; Roth, M.; Zhong, B.; Lan, X.; Tian, J.; He, Q.; Li, D.; Sun, J.; et al. Specific Regulation of PRMT1 Expression by PIAS1 and RKIP in BEAS-2B Epithelia Cells and HFL-1 Fibroblasts in Lung Inflammation. Sci. Rep. 2016, 6, 21810. [Google Scholar] [CrossRef]
- Wright, K.T.; Vella, A.T. RKIP Contributes to IFN-γ Synthesis by CD8+ T Cells after Serial TCR Triggering in Systemic Inflammatory Response Syndrome. J. Immunol. 2013, 191, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-γ–Related mRNA Profile Predicts Clinical Response to PD-1 Blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef]
- Qian, J.; Wang, C.; Wang, B.; Yang, J.; Wang, Y.; Luo, F.; Xu, J.; Zhao, C.; Liu, R.; Chu, Y. The IFN-γ/PD-L1 Axis between T Cells and Tumor Microenvironment: Hints for Glioma Anti-PD-1/PD-L1 Therapy. J. Neuroinflamm. 2018, 15, 290. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Wang, J.; Zhang, J.; Yang, J.; Wang, G.; Wang, Y. Development of Inhibitors Targeting Glycogen Synthase Kinase-3β for Human Diseases: Strategies to Improve Selectivity. Eur. J. Med. Chem. 2022, 236, 114301. [Google Scholar] [CrossRef]
- Al-Mulla, F.; Bitar, M.S.; Al-Maghrebi, M.; Behbehani, A.I.; Al-Ali, W.; Rath, O.; Doyle, B.; Tan, K.Y.; Pitt, A.; Kolch, W. Raf Kinase Inhibitor Protein RKIP Enhances Signaling by Glycogen Synthase Kinase-3β. Cancer Res. 2011, 71, 1334–1343. [Google Scholar] [CrossRef]
- Wu, D.; Pan, W. GSK3: A Multifaceted Kinase in Wnt Signaling. Trends Biochem. Sci. 2010, 35, 161–168. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, Y. Targeting Oncogenic SOX2 in Human Cancer Cells: Therapeutic Application. Protein Cell 2020, 11, 82–84. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.; Cheng, X.; Sun, S.; Zhou, J. Transcriptional Activation of PD-L1 by Sox2 Contributes to the Proliferation of Hepatocellular Carcinoma Cells. Oncol. Rep. 2017, 37, 3061–3067. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.-H.; Park, S.; Kim, S.; Kang, S.-M.; Woo, T.-G.; Yoon, M.-H.; Lee, H.; Jeong, M.; Park, Y.H.; Kim, H.; et al. RKIP Induction Promotes Tumor Differentiation via SOX2 Degradation in NF2-Deficient Conditions. Mol. Cancer Res. 2022, 20, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Ma, D.; Zhang, J.; Zhao, J.; Yang, M. MicroRNA-18a Induces Epithelial-Mesenchymal Transition like Cancer Stem Cell Phenotype via Regulating RKIP Pathway in Pancreatic Cancer. Ann. Transl. Med. 2020, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Wottrich, S.; Bonavida, B. Crosstalks between Raf-Kinase Inhibitor Protein and Cancer Stem Cell Transcription Factors (Oct4, KLF4, Sox2, Nanog). Tumor Biol. 2017, 39, 101042831769225. [Google Scholar] [CrossRef] [PubMed]
- Sarvagalla, S.; Kolapalli, S.P.; Vallabhapurapu, S. The Two Sides of YY1 in Cancer: A Friend and a Foe. Front. Oncol. 2019, 9, 1230. [Google Scholar] [CrossRef] [PubMed]
- Baritaki, S.; Chapman, A.; Yeung, K.; Spandidos, D.A.; Palladino, M.; Bonavida, B. Inhibition of Epithelial to Mesenchymal Transition in Metastatic Prostate Cancer Cells by the Novel Proteasome Inhibitor, NPI-0052: Pivotal Roles of Snail Repression and RKIP Induction. Oncogene 2009, 28, 3573–3585. [Google Scholar] [CrossRef] [PubMed]
- Hays, E.; Bonavida, B. YY1 Regulates Cancer Cell Immune Resistance by Modulating PD-L1 Expression. Drug Resist. Updates 2019, 43, 10–28. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Yan, L.; Liu, N.; Xu, M.; Cai, H. IFI16 Promotes Cervical Cancer Progression by Upregulating PD-L1 in Immunomicroenvironment through STING-TBK1-NF-kB Pathway. Biomed. Pharmacother. 2020, 123, 109790. [Google Scholar] [CrossRef]
- Huang, G.; Wen, Q.; Zhao, Y.; Gao, Q.; Bai, Y. NF-κB Plays a Key Role in Inducing CD274 Expression in Human Monocytes after Lipopolysaccharide Treatment. PLoS ONE 2013, 8, e61602. [Google Scholar] [CrossRef]
- Maeda, T.; Hiraki, M.; Jin, C.; Rajabi, H.; Tagde, A.; Alam, M.; Bouillez, A.; Hu, X.; Suzuki, Y.; Miyo, M.; et al. MUC1-C Induces PD-L1 and Immune Evasion in Triple-Negative Breast Cancer. Cancer Res. 2018, 78, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Gowrishankar, K.; Gunatilake, D.; Gallagher, S.J.; Tiffen, J.; Rizos, H.; Hersey, P. Inducible but Not Constitutive Expression of PD-L1 in Human Melanoma Cells Is Dependent on Activation of NF-κB. PLoS ONE 2015, 10, e0123410. [Google Scholar] [CrossRef] [PubMed]
- Karimi, S.; Mansouri, S.; Mamatjan, Y.; Liu, J.; Nassiri, F.; Suppiah, S.; Singh, O.; Aldape, K.; Zadeh, G. Programmed Death Ligand-1 (PD-L1) Expression in Meningioma; Prognostic Significance and Its Association with Hypoxia and NFKB2 Expression. Sci. Rep. 2020, 10, 14115. [Google Scholar] [CrossRef]
- Li, H.; Xia, J.; Zhu, F.; Xi, Z.; Pan, C.; Gu, L.; Tian, Y. LPS Promotes the Expression of PD-L1 in Gastric Cancer Cells through NF-κB Activation. J. Cell. Biochem. 2018, 119, 9997–10004. [Google Scholar] [CrossRef] [PubMed]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An Integrated Repository Portal for Tumor–Immune System Interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Daily, K.; Patel, V.R.; Rigor, P.; Xie, X.; Baldi, P. MotifMap: Integrative Genome-Wide Maps of Regulatory Motif Sites for Model Species. BMC Bioinform. 2011, 12, 495. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, S.; Bredy, T.; Wood, M.; Spitale, R.C.; Baldi, P. MotifMap-RNA: A Genome-Wide Map of RBP Binding Sites. Bioinformatics 2017, 33, 2029–2031. [Google Scholar] [CrossRef]
- Xie, X.; Rigor, P.; Baldi, P. MotifMap: A Human Genome-Wide Map of Candidate Regulatory Motif Sites. Bioinformatics 2009, 25, 167–174. [Google Scholar] [CrossRef]
- Maniatis, T. The High Mobility Group Protein HMG I(Y) Is Required for NF-KB-Dependent Virus Induction of the Human IFN-P Gene. Cell 1992, 71, 777–789. [Google Scholar]
- Clevers, H. Wnt/β-Catenin Signaling in Development and Disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef]
- King, M.; Murphy, L. American Ginseng (Panax quinquefolius L.) Extract Alters Mitogen-Activated Protein Kinase Cell Signaling and Inhibits Proliferation of MCF-7 Cells. J. Exp. Ther. Oncol. 2007, 6, 147–155. [Google Scholar]
- Kim, S.O.; Kim, M.R. (-)-Epigallocatechin 3-Gallate Inhibits Invasion by Inducing the Expression of Raf Kinase Inhibitor Protein in AsPC-1 Human Pancreatic Adenocarcinoma Cells through the Modulation of Histone Deacetylase Activity. Int. J. Oncol. 2013, 42, 349–358. [Google Scholar] [CrossRef]
- Suhail, M.; Rehan, M.; Tarique, M.; Tabrez, S.; Husain, A.; Zughaibi, T.A. Targeting a Transcription Factor NF-κB by Green Tea Catechins Using in Silico and in Vitro Studies in Pancreatic Cancer. Front. Nutr. 2023, 9, 1078642. [Google Scholar] [CrossRef]
- Jazirehi, A.R.; Bonavida, B. Cellular and Molecular Signal Transduction Pathways Modulated by Rituximab (Rituxan, Anti-CD20 mAb) in Non-Hodgkin’s Lymphoma: Implications in Chemosensitization and Therapeutic Intervention. Oncogene 2005, 24, 2121–2143. [Google Scholar] [CrossRef] [PubMed]
- Singhal, J.; Nagaprashantha, L.D.; Vatsyayan, R.; Ashutosh; Awasthi, S.; Singhal, S.S. Didymin Induces Apoptosis by Inhibiting N-Myc and Upregulating RKIP in Neuroblastoma. Cancer Prev. Res. 2012, 5, 473–483. [Google Scholar] [CrossRef]
- Hu, C.; Zhou, L.; Cai, Y. Dihydroartemisinin Induces Apoptosis of Cervical Cancer Cells via Upregulation of RKIP and Downregulation of Bcl-2. Cancer Biol. Ther. 2014, 15, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Jin, G.; Fujimoto, M. Dihydroartemisinin: A Potential Drug for the Treatment of Malignancies and Inflammatory Diseases. Front. Oncol. 2021, 11, 722331. [Google Scholar] [CrossRef]
- Huang, Q.; Bai, F.; Nie, J.; Lu, S.; Lu, C.; Zhu, X.; Zhuo, L.; Lin, X. Didymin Ameliorates Hepatic Injury through Inhibition of MAPK and NF-κB Pathways by up-Regulating RKIP Expression. Int. Immunopharmacol. 2017, 42, 130–138. [Google Scholar] [CrossRef]
- Cho, J.-H.; Oh, A.-Y.; Park, S.; Kang, S.; Yoon, M.-H.; Woo, T.-G.; Hong, S.-D.; Hwang, J.; Ha, N.-C.; Lee, H.-Y.; et al. Loss of NF2 Induces TGFβ Receptor 1–Mediated Noncanonical and Oncogenic TGFβ Signaling: Implication of the Therapeutic Effect of TGFβ Receptor 1 Inhibitor on NF2 Syndrome. Mol. Cancer Ther. 2018, 17, 2271–2284. [Google Scholar] [CrossRef] [PubMed]
- Bonavida, B.; Garban, H. Nitric Oxide-Mediated Sensitization of Resistant Tumor Cells to Apoptosis by Chemo-Immunotherapeutics. Redox Biol. 2015, 6, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-L.; Chang, M.-C.; Cheng, W.-F. Metronomic Chemotherapy and Immunotherapy in Cancer Treatment. Cancer Lett. 2017, 400, 282–292. [Google Scholar] [CrossRef]
- Huang, M.; Yang, J.; Wang, T.; Song, J.; Xia, J.; Wu, L.; Wang, W.; Wu, Q.; Zhu, Z.; Song, Y.; et al. Homogeneous, Low-volume, Efficient, and Sensitive Quantitation of Circulating Exosomal PD-L1 for Cancer Diagnosis and Immunotherapy Response Prediction. Angew. Chem. 2020, 59, 4800–4805. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Jiang, L.; Li, S.; He, Q.; Yang, B.; Cao, J. Small Molecule Inhibitors Targeting the PD-1/PD-L1 Signaling Pathway. Acta Pharmacol. Sin. 2021, 42, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, S.; Yuan, R.; Engleman, E.G. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu. Rev. Pathol. Mech. Dis. 2021, 16, 223–249. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, Z.; Zheng, X.; Tao, H.; Zhang, S.; Ma, J.; Liu, Z.; Wang, J.; Qian, Y.; Cui, P.; et al. Response Efficacy of PD-1 and PD-L1 Inhibitors in Clinical Trials: A Systematic Review and Meta-Analysis. Front. Oncol. 2021, 11, 562315. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Chen, Z.; Chen, D.; Yan, D. Strategies Targeting PD-L1 Expression and Associated Opportunities for Cancer Combination Therapy. Theranostics 2023, 13, 1520–1544. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Zhang, J.; Li, A.; Niu, M.; Yan, Y.; Jiao, Y.; Luo, S.; Zhou, P.; Wu, K. The Construction, Expression, and Enhanced Anti-Tumor Activity of YM101: A Bispecific Antibody Simultaneously Targeting TGF-β and PD-L1. J. Hematol. Oncol. 2021, 14, 27. [Google Scholar] [CrossRef]
- Wan, Y.Y.; Flavell, R.A. ‘Yin-Yang’ Functions of TGF-β and Tregs in Immune Regulation. Immunol. Rev. 2007, 220, 199–213. [Google Scholar] [CrossRef]
- Melaiu, O.; Mina, M.; Chierici, M.; Boldrini, R.; Jurman, G.; Romania, P.; D’Alicandro, V.; Benedetti, M.C.; Castellano, A.; Liu, T.; et al. PD-L1 Is a Therapeutic Target of the Bromodomain Inhibitor JQ1 and, Combined with HLA Class I, a Promising Prognostic Biomarker in Neuroblastoma. Clin. Cancer Res. 2017, 23, 4462–4472. [Google Scholar] [CrossRef]
- Kim, S.; Koh, J.; Kim, M.-Y.; Kwon, D.; Go, H.; Kim, Y.A.; Jeon, Y.K.; Chung, D.H. PD-L1 Expression Is Associated with Epithelial-to-Mesenchymal Transition in Adenocarcinoma of the Lung. Hum. Pathol. 2016, 58, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Singhal, S.S.; Singhal, S.; Singhal, P.; Singhal, J.; Horne, D.; Awasthi, S. Didymin: An Orally Active Citrus Flavonoid for Targeting Neuroblastoma. Oncotarget 2017, 8, 29428–29441. [Google Scholar] [CrossRef]
- Molina, J.R.; Adjei, A.A. The Ras/Raf/MAPK Pathway. J. Thorac. Oncol. 2006, 1, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Ehrenreiter, K.; Menon, J.; Menard, R.; Kern, F.; Nakazawa, Y.; Bevilacqua, E.; Imamoto, A.; Baccarini, M.; Rosner, M.R. RKIP Regulates MAP Kinase Signaling in Cells with Defective B-Raf Activity. Cell. Signal. 2013, 25, 1156–1165. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Yeung, M.L.; Beach, S.; Shields, J.M.; Yeung, K.C. RKIP Downregulates B-Raf Kinase Activity in Melanoma Cancer Cells. Oncogene 2005, 24, 3535–3540. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Su, F.; Gautam, R.; Wang, N.; Zhang, Y.; Wang, X. Raf Kinase Inhibitor Protein Negatively Regulates FcεRI-Mediated Mast Cell Activation and Allergic Response. Proc. Natl. Acad. Sci. USA 2018, 115, E9859–E9868. [Google Scholar] [CrossRef]
- Lakshmi, S.P.; Reddy, A.T.; Kodidhela, L.D.; Varadacharyulu, N.C. The Tea Catechin Epigallocatechin Gallate Inhibits NF-κB-Mediated Transcriptional Activation by Covalent Modification. Arch. Biochem. Biophys. 2020, 695, 108620. [Google Scholar] [CrossRef] [PubMed]
- Baritaki, S.; Yeung, K.; Palladino, M.; Berenson, J.; Bonavida, B. Pivotal Roles of Snail Inhibition and RKIP Induction by the Proteasome Inhibitor NPI-0052 in Tumor Cell Chemoimmunosensitization. Cancer Res. 2009, 69, 8376–8385. [Google Scholar] [CrossRef]
- Lei, Q.; Wang, D.; Sun, K.; Wang, L.; Zhang, Y. Resistance Mechanisms of Anti-PD1/PDL1 Therapy in Solid Tumors. Front. Cell Dev. Biol. 2020, 8, 672. [Google Scholar] [CrossRef]
- Vivarelli, S.; Falzone, L.; Grillo, C.M.; Bonavida, B.; Crimi, C.; La Mantia, I.; Libra, M. Computational Analyses of YY1 and Its Target RKIP Reveal Their Diagnostic and Prognostic Roles in Lung Cancer. Cancers 2022, 14, 922. [Google Scholar] [CrossRef] [PubMed]
- Atchison, M.L.; Basu, A.; Zaprazna, K.; Papasani, M. Mechanisms of Yin Yang 1 in Oncogenesis: The Importance of Indirect Effects. Crit. Rev. Oncog. 2011, 16, 143–161. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, Z.; Tao, J.; Hu, H.; Li, Z.; Zhang, Z.; Cheng, F.; Sun, Y.; Zhang, Y.; Yang, J.; et al. Resveratrol Induces PD-L1 Expression through Snail-Driven Activation of Wnt Pathway in Lung Cancer Cells. J. Cancer Res. Clin. Oncol. 2021, 147, 1101–1113. [Google Scholar] [CrossRef] [PubMed]
- Noman, M.Z.; Desantis, G.; Janji, B.; Hasmim, M.; Karray, S.; Dessen, P.; Bronte, V.; Chouaib, S. PD-L1 Is a Novel Direct Target of HIF-1α, and Its Blockade under Hypoxia Enhanced MDSC-Mediated T Cell Activation. J. Exp. Med. 2014, 211, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Srivani, G.; Behera, S.K.; Dariya, B.; Chalikonda, G.; Alam, A.; Nagaraju, G.P. HIF-1α and RKIP: A Computational Approach for Pancreatic Cancer Therapy. Mol. Cell. Biochem. 2020, 472, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Vrankar, M.; Kern, I.; Stanic, K. Prognostic Value of PD-L1 Expression in Patients with Unresectable Stage III Non-Small Cell Lung Cancer Treated with Chemoradiotherapy. Radiat. Oncol. 2020, 15, 247. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Tian, M.; Zhang, X.; Qi, C.; Song, C. Distribution of PD-L1 Expression Level across Major Tumor Types. J. Clin. Oncol. 2020, 38, e15176. [Google Scholar] [CrossRef]
- Yuan, J.; Dong, X.; Yap, J.; Hu, J. The MAPK and AMPK Signalings: Interplay and Implication in Targeted Cancer Therapy. J. Hematol. Oncol. 2020, 13, 113. [Google Scholar] [CrossRef]




| RKIP Inducers | Mechanisms | References |
| EGCG | Upregulates RKIP expression by modulating histone deacetylation. Also inhibits Snail and NF-κB activity. | [123,124,125] |
| Didymin | Induces apoptosis by upregulating RKIP and inhibiting N-Myc. | [127] |
| DHA | Induces apoptosis of cervical cancer cells via upregulation of RKIP in HeLa cells. | [128] |
| Nf18001 | Inhibitions of the RKIP– TβR1 network. | [131] |
| DETA/NO | Enhances RKIP expression by inhibiting the NF-κB/YY1/Snail regulatory circuit. | [75,132] |
| NPI-0052 | Proteasome inhibitor. | [108] |
| Ginseng Extract | Correlations with a significant increase in RKIP mRNA and protein expression. | [123] |
| PD-L1 mAbs | Mechanism | References |
| Aavelumab | Binds to PD-L1, preventing the interaction between PD-L1 and PD-1. | [50,51,52] |
| Atezolizumab | Binds to PD-L1, preventing the interaction between PD-L1 and PD-1. | [50,51,52] |
| Durvalumab | Binds to PD-L1, preventing the interaction between PD-L1 and PD-1. | [50,51,52] |
| Type of Malignancy | Phase | Interventions | Relation to RKIP | NCT Reference |
|---|---|---|---|---|
| Melanoma | 2 |
Pembrolizumab Dabrafenib Trametinib | Trial targets the MAPK pathway using ERK and BRAF inhibitors. RKIP can inhibit MAPK/ERK signaling via Raf-1 [139]. RKIP expression can also inhibit BRAF in melanoma cell line [141]. | NCT03149029 |
| Melanoma | 1 |
Durvalumab Dabrafenib Trametinib | Trial targets the MAPK pathway using ERK and BRAF inhibitors. RKIP can inhibit MAPK/ERK signaling via Raf-1 [139]. RKIP expression can also inhibit BRAF in melanoma cell line [141]. | NCT02027961 |
| Refractory Melanoma and Other Malignant Neoplasms of Skin | 1/2 | GSK2636771 (PI3K-Beta Inhibitor) Pembrolizumab | Trial targets the PI3K pathway. RKIP interacts with PI3K, preventing it from binding to GRB2-associated binding protein 2 (Gab2) [142]. | NCT03131908 |
| Sarcomas Including Malignant Peripheral Nerve Sheath Tumors | 1/2 |
Selumetinib and Bromodomain (MEK inhibitors) Durvalumab | Trial targets the MAPK pathway. RKIP inhibits MAPK and MEK signaling via Raf-1 [139]. | NCT05253131 |
| Type of Malignancy | Phase | Interventions | Relation to RKIP | NCT Reference |
|---|---|---|---|---|
| Solid Tumors, Lymphomas, Leukemias, and Multiple Myeloma | 1 |
NPI-0052, Dexamethasone | NPI-0052 has been shown to induce RKIP [108]. | NCT00629473 |
| Grade IV Malignant Glioma | 1 | NPI-0052, Radiotherapy (RT), Temozolomide (TMZ), and Optune | NPI-0052 has been shown to induce RKIP [108]. | NCT02903069 |
| Glioblastoma | 3 | NPI-0052, RT, TMZ | NPI-0052 has been shown to induce RKIP [108]. | NCT03345095 |
| Multiple Myeloma | 2 | NPI-0052 | NPI-0052 has been shown to induce RKIP [108]. | NCT00461045 |
| Prostate Cancer | 2 | EGCG | EGCG has been shown to induce RKIP [124]. | NCT00676780 |
| Breast Cancer | 2 | EGCG | EGCG has been shown to induce RKIP [124]. | NCT00917735 |
| Bladder Cancer | 2 | EGCG | EGCG has been shown to induce RKIP [124]. | NCT00666562 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ho, M.; Bonavida, B. Cross-Talks between Raf Kinase Inhibitor Protein and Programmed Cell Death Ligand 1 Expressions in Cancer: Role in Immune Evasion and Therapeutic Implications. Cells 2024, 13, 864. https://doi.org/10.3390/cells13100864
Ho M, Bonavida B. Cross-Talks between Raf Kinase Inhibitor Protein and Programmed Cell Death Ligand 1 Expressions in Cancer: Role in Immune Evasion and Therapeutic Implications. Cells. 2024; 13(10):864. https://doi.org/10.3390/cells13100864
Chicago/Turabian StyleHo, Mai, and Benjamin Bonavida. 2024. "Cross-Talks between Raf Kinase Inhibitor Protein and Programmed Cell Death Ligand 1 Expressions in Cancer: Role in Immune Evasion and Therapeutic Implications" Cells 13, no. 10: 864. https://doi.org/10.3390/cells13100864





