Morphological and Molecular Analysis Identified a Subspecies of Crassostrea ariakensis (Fujita, 1913) along the Coast of Asia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples and Data Collection
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Phylogenetic Analysis
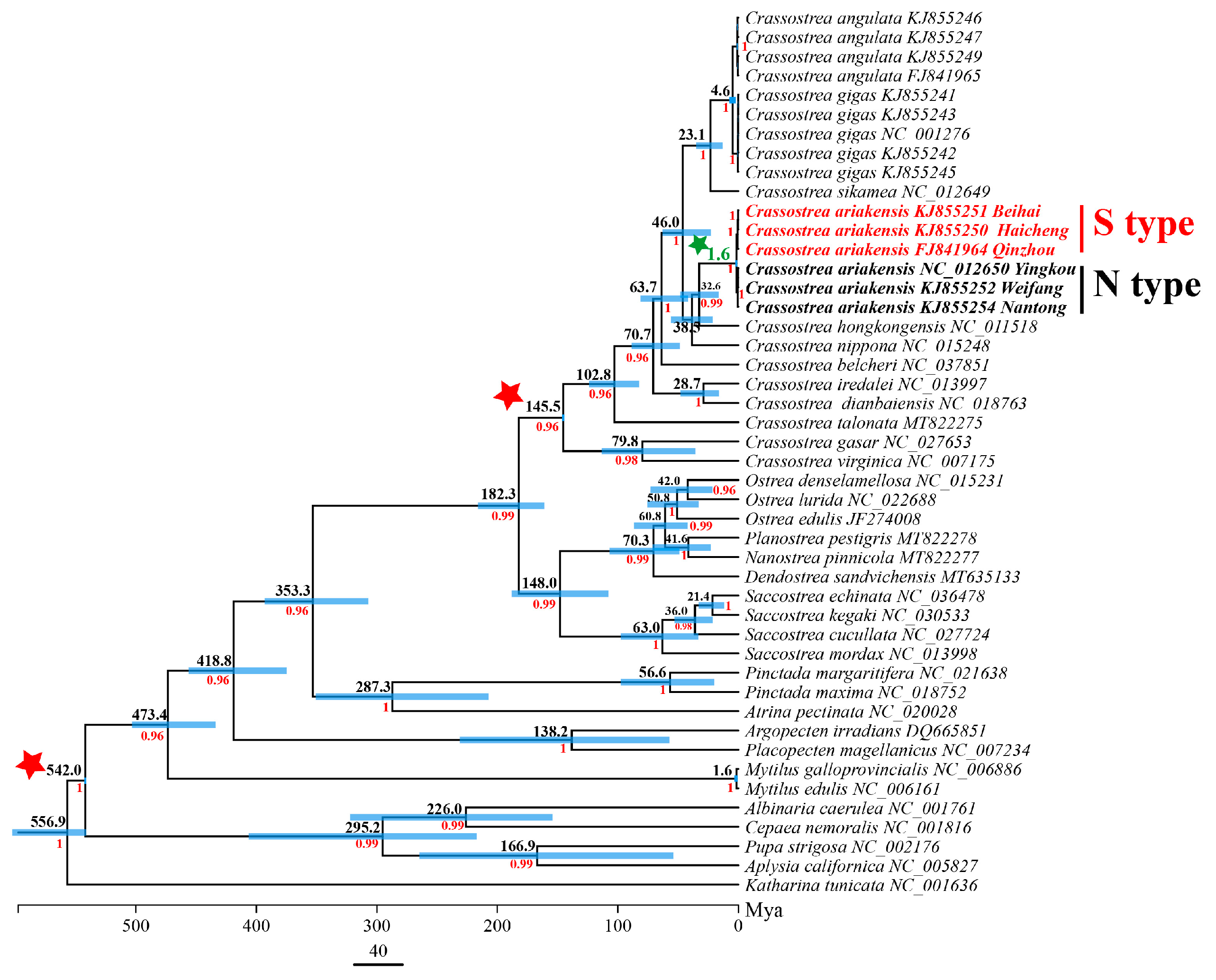
2.4. Divergence Time Estimation
2.5. Molecular Species Delimitation Analysis
3. Results
3.1. Shell Morphology
3.2. COI Sequences
3.3. 16S rRNA Sequences
3.4. Mitogenome Sequences and Divergence Time Estimation
3.5. Species Delimitation Analysis
4. Discussion
4.1. Identification of C. ariakensis Subspecies
- Systematics
4.2. Distribution of C. ariakensis Subspecies
4.3. Relationship between C. ariakensis ariakensis subsp. nov. and C. ariakensis meridioyangtzensis subsp. nov.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| SR | SA | KD | AKB | IR | YK | BZ | DY | GR | WF | NT | SH | HY | FH | XM | ST | SZ | ZH | HMT | CDZ | YJ | ZJ | BH | QZ | FCG | HK | VN | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap1 | 5 | 1 | 1 | 33 | 2 | 2 | 5 | 8 | 31 | 2 | 28 | 4 | 13 | 4 | |||||||||||||
| Hap2 | 1 | 1 | 1 | ||||||||||||||||||||||||
| Hap3 | 1 | 1 | |||||||||||||||||||||||||
| Hap4 | 1 | ||||||||||||||||||||||||||
| Hap5 | 1 | ||||||||||||||||||||||||||
| Hap6 | 1 | 1 | |||||||||||||||||||||||||
| Hap7 | 1 | ||||||||||||||||||||||||||
| Hap8 | 1 | ||||||||||||||||||||||||||
| Hap9 | 1 | ||||||||||||||||||||||||||
| Hap10 | 1 | ||||||||||||||||||||||||||
| Hap11 | 28 | 12 | 2 | 10 | 1 | 1 | 1 | 27 | 1 | 8 | 17 | 1 | |||||||||||||||
| Hap12 | 1 | 2 | |||||||||||||||||||||||||
| Hap13 | 2 | ||||||||||||||||||||||||||
| Hap14 | 1 | ||||||||||||||||||||||||||
| Hap15 | 1 | ||||||||||||||||||||||||||
| Hap16 | 1 | ||||||||||||||||||||||||||
| Hap17 | 1 | ||||||||||||||||||||||||||
| Hap18 | 2 | ||||||||||||||||||||||||||
| Hap19 | 1 | ||||||||||||||||||||||||||
| Hap20 | 1 | ||||||||||||||||||||||||||
| Hap21 | 1 | ||||||||||||||||||||||||||
| Hap22 | 1 | ||||||||||||||||||||||||||
| Hap23 | 1 | ||||||||||||||||||||||||||
| Hap24 | 1 | ||||||||||||||||||||||||||
| Hap25 | 1 | ||||||||||||||||||||||||||
| Hap26 | 1 | 1 | |||||||||||||||||||||||||
| Hap27 | 1 | ||||||||||||||||||||||||||
| Hap28 | 1 | ||||||||||||||||||||||||||
| Hap29 | 1 | ||||||||||||||||||||||||||
| Hap30 | 1 | ||||||||||||||||||||||||||
| Hap31 | 1 | ||||||||||||||||||||||||||
| Hap32 | 1 | ||||||||||||||||||||||||||
| Hap33 | 1 | ||||||||||||||||||||||||||
| Hap34 | 1 | ||||||||||||||||||||||||||
| Hap35 | 1 | 2 | |||||||||||||||||||||||||
| Hap36 | 1 | ||||||||||||||||||||||||||
| Hap37 | 1 | ||||||||||||||||||||||||||
| Hap38 | 1 | ||||||||||||||||||||||||||
| Hap39 | 1 | ||||||||||||||||||||||||||
| Hap40 | 1 | 1 | |||||||||||||||||||||||||
| Hap41 | 1 | ||||||||||||||||||||||||||
| Hap42 | 2 | ||||||||||||||||||||||||||
| Hap43 | 1 | ||||||||||||||||||||||||||
| Hap44 | 1 | ||||||||||||||||||||||||||
| Hap45 | 1 | ||||||||||||||||||||||||||
| Hap46 | 1 | ||||||||||||||||||||||||||
| Hap47 | 1 | ||||||||||||||||||||||||||
| Hap48 | 1 | ||||||||||||||||||||||||||
| Hap49 | 2 | ||||||||||||||||||||||||||
| Hap50 | 1 | ||||||||||||||||||||||||||
| Hap51 | 1 | ||||||||||||||||||||||||||
| Hap52 | 1 | ||||||||||||||||||||||||||
| Hap53 | 1 | ||||||||||||||||||||||||||
| Hap54 | 1 | ||||||||||||||||||||||||||
| Hap55 | 2 | ||||||||||||||||||||||||||
| Hap56 | 2 | ||||||||||||||||||||||||||
| Hap57 | 1 | 8 | 6 | ||||||||||||||||||||||||
| Hap58 | 1 | ||||||||||||||||||||||||||
| Hap59 | 1 | ||||||||||||||||||||||||||
| Hap60 | 1 | ||||||||||||||||||||||||||
| Hap61 | 1 | ||||||||||||||||||||||||||
| Hap62 | 1 | ||||||||||||||||||||||||||
| Hap63 | 1 | ||||||||||||||||||||||||||
| Hap64 | 1 | ||||||||||||||||||||||||||
| Hap65 | 1 | ||||||||||||||||||||||||||
| Hap66 | 1 |
| SR | SA | KD | IR | YK | BZ | DY | WF | NT | SH | HY | FH | XM | SZ | HMT | CDZ | YJ | ZJ | BH | QZ | HKQK | VN | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap1 | 6 | 2 | 1 | 2 | 2 | 5 | 10 | 3 | 7 | 5 | 15 | 1 | ||||||||||
| Hap2 | 1 | |||||||||||||||||||||
| Hap3 | 1 | |||||||||||||||||||||
| Hap4 | 4 | 2 | 6 | 2 | 3 | 1 | 1 | 13 | 1 | 1 | 3 | |||||||||||
| Hap5 | 1 | |||||||||||||||||||||
| Hap6 | 1 | |||||||||||||||||||||
| Hap7 | 1 | |||||||||||||||||||||
| Hap8 | 1 | |||||||||||||||||||||
| Hap9 | 1 | |||||||||||||||||||||
| Hap10 | 1 | |||||||||||||||||||||
| Hap11 | 1 | |||||||||||||||||||||
| Hap12 | 1 | |||||||||||||||||||||
| Hap13 | 2 | |||||||||||||||||||||
| Hap14 | 1 | 1 | ||||||||||||||||||||
| Hap15 | 1 | |||||||||||||||||||||
| Hap16 | 1 |
References
- Guo, X.; Li, C.; Wang, H.; Xu, Z. Diversity and Evolution of Living Oysters. J. Shellfish Res. 2018, 37, 755–771. [Google Scholar] [CrossRef]
- Xia, J.; Wu, X.; Xiao, S.; Yu, Z. Mitochondrial DNA and Morphological Identification of a New Cupped Oyster Species Crassostrea Dianbaiensis (Bivalvia: Ostreidae) in the South China Sea. Aquat. Living Resour. 2014, 27, 41–48. [Google Scholar] [CrossRef]
- Wang, H.; Guo, X.; Liu, X.; Zhang, G.; Zhang, S.; Xu, F. Classification of “Jin jiang” oysters in China. Mar. Sci. 2007, 31, 85–86. [Google Scholar]
- Xiao, J.; Cordes, J.F.; Wang, H.; Guo, X.; Reece, K.S. Population Genetics of Crassostrea Ariakensis in Asia Inferred from Microsatellite Markers. Mar. Biol. 2010, 157, 1767–1781. [Google Scholar] [CrossRef]
- Wang, H.; Guo, X.; Zhang, G.; Zhang, F. Classification of Jinjiang Oysters Crassostrea Rivularis (Gould, 1861) from China, Based on Morphology and Phylogenetic Analysis. Aquaculture 2004, 242, 137–155. [Google Scholar] [CrossRef]
- Kim, W.-J.; Dammannagoda, S.T.; Jung, H.; Baek, I.S.; Yoon, H.S.; Choi, S.D. Mitochondrial DNA Sequence Analysis from Multiple Gene Fragments Reveals Genetic Heterogeneity of Crassostrea ariakensis in East Asia. Genes Genom. 2014, 36, 611–624. [Google Scholar] [CrossRef]
- Ren, J.; Hou, Z.; Wang, H.; Sun, M.; Liu, X.; Liu, B.; Guo, X. Intraspecific Variation in Mitogenomes of Five Crassostrea Species Provides Insight into Oyster Diversification and Speciation. Mar. Biotechnol. 2016, 18, 242–254. [Google Scholar] [CrossRef]
- Li, A.; Dai, H.; Guo, X.; Zhang, Z.; Zhang, K.; Wang, C.; Wang, X.; Wang, W.; Chen, H.; Li, X.; et al. Genome of the Estuarine Oyster Provides Insights into Climate Impact and Adaptive Plasticity. Commun. Biol. 2021, 4, 1287. [Google Scholar] [CrossRef]
- Wu, B.; Chen, X.; Yu, M.; Ren, J.; Hu, J.; Shao, C.; Zhou, L.; Sun, X.; Yu, T.; Zheng, Y.; et al. Chromosome-level Genome and Population Genomic Analysis Provide Insights into the Evolution and Environmental Adaptation of Jinjiang Oyster Crassostrea Ariakensis. Mol. Ecol. Resour. 2022, 22, 1529–1544. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Q.; Kong, L.; Yu, H.; Zheng, X. Identifying the True Oysters (Bivalvia: Ostreidae) with Mitochondrial Phylogeny and Distance-based DNA Barcoding. Mol. Ecol. Resour. 2011, 11, 820–830. [Google Scholar] [CrossRef]
- Li, A.; Wang, C.; Wang, W.; Zhang, Z.; Liu, M.; She, Z.; Jia, Z.; Zhang, G.; Li, L. Molecular and Fitness Data Reveal Local Adaptation of Southern and Northern Estuarine Oysters (Crassostrea Ariakensis). Front. Mar. Sci. 2020, 7, 589099. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble Species by Automatic Partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A General Species Delimitation Method with Applications to Phylogenetic Placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Palumbi, S.R.; Martin, A.; Romano, S.; McMillan, W.O.; Stice, L.; Grabowski, G. The Simple Fool’s Guide to PCR; University of Hawaii Press: Honolulu, HI, USA, 1996. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Peterson, K.J.; Cotton, J.A.; Gehling, J.G.; Pisani, D. The Ediacaran Emergence of Bilaterians: Congruence between the Genetic and the Geological Fossil Records. Phil. Trans. R. Soc. B 2008, 363, 1435–1443. [Google Scholar] [CrossRef] [PubMed]
- Li, C. Phylogeny and Evolutionary Analysis of Oysters (Bivalvia: Ostreoidea) and Complete Mitochondrial DNA of Talonostrea talonata. Master’s Thesis, Institution of Oceanology, Chinese Academy of Sciences, Qingdao, China, 2013. Available online: http://ir.qdio.ac.cn/handle/337002/17970 (accessed on 10 March 2024).
- Helfrich, P.; Rieb, E.; Abrami, G. TREEANNOTATOR: Versatile Visual Annotation of Hierarchical Text Relations. In Proceedings of the Eleventh International Conference on Language Resources and Evaluation (LREC 2018), Miyazaki, Japan, 7–12 May 2018; European Language Resources Association (ELRA): Paris, France, 2018. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Wang, H.; Qian, L.; Liu, X.; Zhang, G.; Guo, X. Classification of a Common Cupped Oyster from Southern China. J. Shellfish Res. 2010, 29, 857–866. [Google Scholar] [CrossRef]
- Wang, H. Studies on the Molecular Phylogeny and Taxonomy of Common Oysters in China Seas. Ph.D. Thesis, Institution of Oceanology, Chinese Academy of Sciences, Qingdao, China, 2004. [Google Scholar]
- Xu, F.; Zhang, S. An Illustrated Bivalvia Mollusca Fauna of China Seas, 1st ed.; Science Press: Beijing, China, 2008. [Google Scholar]
- Li, S. Population Genetics Studies on Four Species of Crassostrea Oysters. Master’s Thesis, Ocean University of China, Qingdao, China, 2015. [Google Scholar]
- Ni, G. Phylogeography of Four Marine Bivalves along China’ Coastline, with Views into the Evolutionary Processes and Mechanisms. Ph.D. Thesis, Ocean University of China, Qingdao, China, 2013. [Google Scholar]







| Location | n | Longitude [Degrees East] | Latitude [Degrees North] |
|---|---|---|---|
| SR: Seomjin River, South Korea * | 10 | 121.76 | 34.92 |
| SA: Sacheon Kawha River, South Korea * | 5 | 128.04 | 35.02 |
| KD: Kangwha-do, South Korea * | 3 | 126.35 | 37.74 |
| AKB: Ariake Bay, Japan * | 35 | 130.52 | 32.88 |
| IR: Itoki River, Japan * | 3 | 130.18 | 33.02 |
| YK: Yingkou, Liaonning, China * | 2 | 122.15 | 40.69 |
| BZ: Binzhou, Shandong, China | 5 | 117.85 | 38.25 |
| DY: Kenli, Dongying, Shandong, China | 11 | 119.24 | 37.83 |
| GR: Guangrao, Dongying, Shandong, China * | 36 | 118.94 | 37.35 |
| WF: Weifang, Shandong, China | 6 | 119.05 | 37.29 |
| NT: Nantong, Jiangsu, China | 39 | 121.52 | 32.11 |
| SH: Shanghai, China | 8 | 121.97 | 30.89 |
| HY: Haiyan, Zhejiang, China | 17 | 120.98 | 30.53 |
| FH: Fenghua, Zhejiang, China | 5 | 121.49 | 29.50 |
| XM: Xiamen, Fujian, China * | 34 | 118.19 | 24.66 |
| ST: Shantou, Guangdong, China | 13 | 116.72 | 23.33 |
| SZ: Shenzhen, Guangdong, China | 14 | 114.04 | 22.50 |
| ZH: Zhuhai, Guangdong, China * | 11 | 113.58 | 22.29 |
| HMT: Huangmaotian, Taishan, Guangdong, China | 3 | 113.02 | 21.94 |
| CDZ: Chuandaozhen, Taishan, Guangdong, China | 3 | 112.65 | 21.80 |
| YJ: Yangjiang, Guangdong, China | 2 | 111.85 | 21.66 |
| ZJ: Zhanjiang, Guangdong, China | 39 | 110.43 | 21.21 |
| BH: Beihai, Guangxi, China | 14 | 109.16 | 21.51 |
| QZ: Maoweihai, Qinzhou, Guangxi, China * | 10 | 108.58 | 21.74 |
| FCG: Fangchenggang, Guangxi, China * | 19 | 108.34 | 21.69 |
| HK: Pearl River Delta, Hong Kong, China * | 3 | 114.16 | 22.29 |
| HKQK: Qukou, Haikou, China | 1 | 110.59 | 19.95 |
| VT: Vietnam | 3 | 106.60 | 20.24 |
| Species | GenBank Accession Number | |
|---|---|---|
| COI | 16S rRNA | |
| C.ariakensis | FJ743512-27 KP734018-62 | KX345399-410 FJ743503-07 |
| EU007496-98 | LC005447 | |
| EU007503-05 | EU672835 * NC_012650 * | |
| EU672835 * NC_012650 * | AY632546-48 KJ855250-52, KJ855254 | |
| KX345411-28 | HQ660979-80 KC847118 | |
| AY632559-66 | FJ841964 * | |
| HQ661020-21 | ||
| EU007493 FJ841964 * AY160752-54 | ||
| C.ariakensis | AF300617, KP734060 | AY160757 |
| C. hongkongensis (Lam and B. Morton, 2003) | AJ553912, KP976208 | AY160756, KX345688 |
| C. gigas angulata | AJ553908, AJ553907, KP216805 | AJ553901, AJ553902, KX345694 |
| C. gigas gigas | AF152565, AJ553910, KP099016 | AJ553903, AJ553905, KX345700 |
| C. sikamea (Amemiya, 1928) | AF152568, AB904878 | AY632551, KX345717 |
| C.virginica | AF152566, KU905937 | AF092285 |
| C. rhizophorae (Guilding, 1828) | KP455050 | AJ312938 |
| C. belcheri (G. B. Sowerby II, 1871) | AY160755 | AY160758 |
| C. iredalei (Faustino, 1932) | AY038078 | AJ553913 |
| C. nippona (Seki, 1934) | -- | LC005446 |
| Saccostrea commercialis (Iredale and Roughley, 1933) | -- | AF353100 |
| O. edulis | AF540599 | AF052068 |
| S. cuccullata (Born, 1778) | AY038076 | -- |
| S. glomerata (A. Gould, 1850) | -- | AF353101 |
| Taxon | Species | GenBank Accession Number |
|---|---|---|
| Bivalvia | C. gigas angulata | KJ855247 |
| C. gigas angulata | KJ855249 | |
| C. gigas angulata | KJ855246 | |
| C. gigas angulata | FJ841965 | |
| C. gigas gigas | NC_001276 | |
| C. gigas gigas | KJ855243 | |
| C. gigas gigas | KJ855245 | |
| C. gigas gigas | KJ855242 | |
| C. gigas gigas | KJ855241 | |
| C. sikamea | NC_012649 | |
| C.ariakensis | NC012650 | |
| C.ariakensis | KJ855252 | |
| C.ariakensis | KJ855254 | |
| C.ariakensis | KJ855250 | |
| C.ariakensis | KJ855251 | |
| C.ariakensis | FJ841964 | |
| C. hongkongensis | NC_011518 | |
| C. nippona | NC_015248 | |
| C. becheri | NC_037851 | |
| C. iredalei | NC_013997 | |
| C. dianbaiensis J.-J. Xia, X.-Y. Wu, S. Xiao, and Z. Yu, 2014 | NC_018763 | |
| C. talonata X.-X. Li and Z.-Y. Qi, 1994 | MT822275 | |
| C. gasar (Dautzenberg, 1891) | NC_027653 | |
| C. virginica | NC_007175 | |
| Nanostrea exigua pinnicola (Pagenstecher, 1877) | MT822277 | |
| Planostrea pestigris (Hanley, 1846) | MT822278 | |
| Dendostrea sandvichensis (G. B. Sowerby II, 1871) | MT635133 | |
| O. denselamellosa Lischke, 1869 | NC_015231 | |
| O. edulis | JF274008 | |
| O. lurida P. P. Carpenter, 1864 | NC_022688 | |
| S. echinata (Quoy and Gaimard, 1835) | NC_036478 | |
| S. kegaki Torigoe and Inaba, 1981 | NC_030533 | |
| S. cucullata | NC_027724 | |
| S. mordax (Gould, 1850) | NC_013998 | |
| Pinctada maxima (Jameson, 1901) | NC_018752 | |
| P. margaritifera (Linnaeus, 1758) | NC_021638 | |
| Atrina pectinate (Linnaeus, 1767) | NC_020028 | |
| Mytilus edulis Linnaeus, 1758 | NC_006161 | |
| M. galloprovincialis Lamarck, 1819) | NC_006886 | |
| Argopecten irradians (Lamarck, 1819) | DQ665851 | |
| Placopecten magellanicus (Gmelin, 1791) | NC_007234 | |
| Gastropoda | Albinaria caerulea (Deshayes, 1835) | NC_001761 |
| Aplysia californica J. G. Cooper, 1863 | NC_005827 | |
| Cepaea nemoralis (Linnaeus, 1758) | NC_001816 | |
| Pupa strigosa (A. Gould, 1859) | NC_002176 | |
| Polyplacphora | K. tunicata | NC_001636 |
| Species | Car_N | Car_S | Can | Cgi | Csi | Cvi | Cbe | Cir | Chk | Oed | Scu | Crh |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Car_N | 0.002 | 0.021 | 0.021 | 0.021 | 0.029 | 0.023 | 0.022 | 0.021 | 0.029 | 0.034 | 0.030 | |
| Car_S | 0.006 | 0.021 | 0.021 | 0.020 | 0.030 | 0.024 | 0.022 | 0.020 | 0.029 | 0.033 | 0.029 | |
| Can | 0.164 | 0.161 | 0.007 | 0.016 | 0.025 | 0.023 | 0.022 | 0.019 | 0.031 | 0.033 | 0.027 | |
| Cgi | 0.159 | 0.156 | 0.026 | 0.016 | 0.025 | 0.023 | 0.021 | 0.019 | 0.030 | 0.033 | 0.027 | |
| Csi | 0.164 | 0.161 | 0.105 | 0.114 | 0.026 | 0.022 | 0.023 | 0.019 | 0.029 | 0.032 | 0.027 | |
| Cvi | 0.280 | 0.283 | 0.235 | 0.238 | 0.239 | 0.028 | 0.028 | 0.029 | 0.030 | 0.034 | 0.023 | |
| Cbe | 0.199 | 0.202 | 0.189 | 0.188 | 0.171 | 0.256 | 0.022 | 0.024 | 0.029 | 0.032 | 0.029 | |
| Cir | 0.175 | 0.177 | 0.175 | 0.173 | 0.194 | 0.255 | 0.185 | 0.022 | 0.029 | 0.030 | 0.030 | |
| Chk | 0.152 | 0.148 | 0.138 | 0.137 | 0.147 | 0.260 | 0.203 | 0.166 | 0.030 | 0.034 | 0.029 | |
| Oed | 0.292 | 0.289 | 0.300 | 0.292 | 0.271 | 0.293 | 0.278 | 0.262 | 0.291 | 0.029 | 0.031 | |
| Scu | 0.324 | 0.321 | 0.310 | 0.306 | 0.312 | 0.347 | 0.318 | 0.276 | 0.323 | 0.269 | 0.035 | |
| Crh | 0.282 | 0.279 | 0.256 | 0.259 | 0.254 | 0.183 | 0.274 | 0.285 | 0.275 | 0.310 | 0.352 |
| Species | Car_N | Car_S | Can | Cgi | Csi | Cvi | Crh | Cbe | Cir | Chk | Sco | Oed | Sgl | Cni |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Car_N | 0.003 | 0.012 | 0.012 | 0.011 | 0.030 | 0.024 | 0.013 | 0.012 | 0.010 | 0.025 | 0.022 | 0.025 | 0.014 | |
| Car_S | 0.006 | 0.011 | 0.012 | 0.011 | 0.029 | 0.024 | 0.012 | 0.012 | 0.010 | 0.025 | 0.022 | 0.025 | 0.014 | |
| Can | 0.053 | 0.051 | 0.004 | 0.007 | 0.029 | 0.023 | 0.011 | 0.011 | 0.007 | 0.026 | 0.023 | 0.026 | 0.011 | |
| Cgi | 0.056 | 0.055 | 0.008 | 0.007 | 0.029 | 0.023 | 0.012 | 0.011 | 0.008 | 0.027 | 0.023 | 0.027 | 0.011 | |
| Csi | 0.050 | 0.047 | 0.018 | 0.022 | 0.030 | 0.023 | 0.010 | 0.010 | 0.008 | 0.025 | 0.023 | 0.025 | 0.012 | |
| Cvi | 0.272 | 0.270 | 0.264 | 0.262 | 0.272 | 0.025 | 0.030 | 0.032 | 0.030 | 0.039 | 0.035 | 0.039 | 0.030 | |
| Crh | 0.190 | 0.187 | 0.173 | 0.173 | 0.176 | 0.209 | 0.024 | 0.023 | 0.024 | 0.032 | 0.029 | 0.032 | 0.026 | |
| Cbe | 0.059 | 0.056 | 0.048 | 0.056 | 0.044 | 0.284 | 0.190 | 0.010 | 0.010 | 0.027 | 0.024 | 0.027 | 0.013 | |
| Cir | 0.059 | 0.060 | 0.047 | 0.050 | 0.038 | 0.299 | 0.183 | 0.041 | 0.010 | 0.027 | 0.024 | 0.027 | 0.012 | |
| Chk | 0.036 | 0.040 | 0.022 | 0.025 | 0.024 | 0.272 | 0.179 | 0.041 | 0.038 | 0.026 | 0.022 | 0.026 | 0.010 | |
| Sco | 0.203 | 0.201 | 0.212 | 0.214 | 0.202 | 0.392 | 0.299 | 0.227 | 0.216 | 0.210 | 0.028 | 0.000 | 0.027 | |
| Oed | 0.166 | 0.163 | 0.182 | 0.183 | 0.172 | 0.338 | 0.241 | 0.182 | 0.182 | 0.175 | 0.245 | 0.028 | 0.026 | |
| Sgl | 0.203 | 0.201 | 0.212 | 0.214 | 0.202 | 0.392 | 0.299 | 0.227 | 0.216 | 0.210 | 0.000 | 0.245 | 0.027 | |
| Cni | 0.072 | 0.076 | 0.046 | 0.045 | 0.050 | 0.276 | 0.203 | 0.067 | 0.064 | 0.041 | 0.228 | 0.203 | 0.228 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Li, C.; Lu, R.; Wang, H. Morphological and Molecular Analysis Identified a Subspecies of Crassostrea ariakensis (Fujita, 1913) along the Coast of Asia. Genes 2024, 15, 644. https://doi.org/10.3390/genes15050644
Chen Y, Li C, Lu R, Wang H. Morphological and Molecular Analysis Identified a Subspecies of Crassostrea ariakensis (Fujita, 1913) along the Coast of Asia. Genes. 2024; 15(5):644. https://doi.org/10.3390/genes15050644
Chicago/Turabian StyleChen, Ya, Cui Li, Ruijing Lu, and Haiyan Wang. 2024. "Morphological and Molecular Analysis Identified a Subspecies of Crassostrea ariakensis (Fujita, 1913) along the Coast of Asia" Genes 15, no. 5: 644. https://doi.org/10.3390/genes15050644





