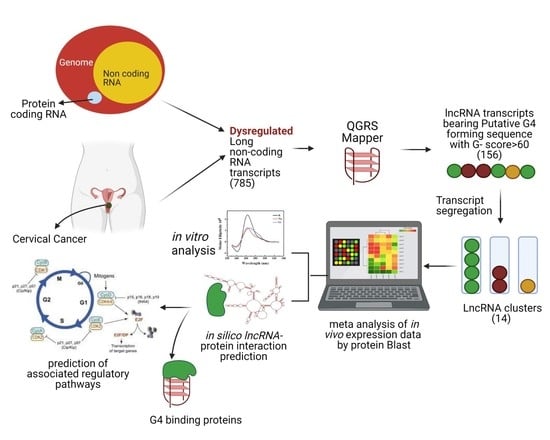
1. Introduction
Non-coding RNA (ncRNA), as the name suggests, are the RNAs that do not code for any protein. These sequences outnumber protein-coding sequences in the human genome. These nucleic acids were once considered dark matter and rendered unimportant due to their perceived disconnect from the central dogma [
1]. However, non-coding RNAs have now assumed prominence for their gene-regulation roles. The largest group of ncRNAs includes transcripts that are over 200 nucleotides long and are termed long non-coding RNAs (lncRNAs). The spatiotemporal expression of lncRNAs across cell types has been correlated with several key cellular functions such as replication, transcription, translation, immune response, angiogenesis and apoptosis [
2]. Further, the dysregulated expression of lncRNA transcripts has been correlated with various pathological conditions including cancer [
3]. Cancer is a complex disease that alters the genomic and proteomic homeostasis of the cell to promote growth and proliferation [
4]. The identification of specific biomarkers has revolutionized the early detection of cancers. Many cancers are curable if diagnosed at an early stage followed by suitable and timely treatment [
5]. Nevertheless, cervical cancer causes the second highest number of deaths among women in India [
6]. This is an alarming statistic considering that most cervical cancers can be successfully treated and human papillomavirus (HPV)-induced cancer can be prevented by vaccines [
7]. The role of lncRNAs in tumorigenesis, metastasis and radio-resistance has compelled researchers to study them as potential cancer biomarkers [
8]. While the precise mechanism by which lncRNAs control cancer dynamics is largely unknown, regulatory lncRNAs can serve as biomarkers of malignancies in different cancer phenotypes [
9]. LncRNAs are notable for their heterogeneity, with sequence conservation across species ranging from very high to none. Moreover, sequence conservation does not guarantee functional resemblance in lncRNAs. Therefore, it is more intuitive to postulate a structure–function relationship for lncRNAs which allows them to access multiple binding sites for proteins, miRNA, mRNA, etc. [
10]. LncRNAs range in length from a few hundred to several thousand nucleotides, folding into a plethora of complex secondary structures including G4s. rG4 structures are stabilized by K
+ ions. The abundance of K
+ ions inside human cells likely facilitates the adoption of G4 structures by RNA in relation to other RNA secondary structures [
11,
12]. Nevertheless, G4s have been suggested to maintain a dynamic equilibrium in vitro, between unfolded and folded states. Such equilibria appear likely in vivo with favorable intracellular K
+ concentrations and the presence of helicases capable of resolving the structures. The G4RP-seq technique developed by Yang et al. supports the existence of transient G4-RNA in the human transcriptome. The study reported that lncRNAs avoid G4 formation under normal conditions in the absence of G4-binding ligands and when a lncRNA such as Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) folds spontaneously into G4, then it is immediately countered or resolved by helicases and RNA-binding proteins (RBPs) [
13]. Many reports have emerged that suggest implications of G-quadruplexes in key cancer-linked lncRNAs [
9,
12]. G-Quadruplex Forming Sequence Containing LncRNA (GSEC) was one of the first lncRNAs identified bearing a G-quadruplex structure and its importance in GSEC-mediated colorectal cancer cell migration was elucidated [
14]. LINC00273, LncRNA In Non-Homologous End Joining Pathway 1 (LINP1), Nuclear Paraspeckle Assembly Transcript 1 (NEAT1), and Lung Cancer-Associated Transcript 1 (LUCAT1) are examples of other lncRNAs which are proposed biomarkers in different types of cancer and that execute their function via G4 secondary structures [
15,
16,
17,
18].
The in silico identification of G4s in lncRNAs is challenging because RNA folding/structure-prediction algorithms do not explicitly account for putative G4 sequences. The Vienna RNA folding suite estimates RNA G4 folding energy and assesses the competition between G4 folded and alternative RNA secondary structures [
19]. However, there is a limitation to sequence input in such predictive tools, and the fact that lncRNAs are up to several thousand nucleotides long cannot be accepted as query sequences. In this work, we present a workflow that enables in silico identification of potential quadruplex-forming sequences in lncRNAs of cervical cancer. Subsequent in vitro analysis validates the G4-forming potential of our present in silico lncRNA predictions. As part of our in silico pipeline, we present two approaches to predict protein-interacting partners of cognate lncRNAs. We have strategically deployed several tools and databases with the goal of recognizing G4-forming lncRNAs in cervical cancer with potential prognostic capabilities. The overall workflow in the present work is illustrated in the graphical abstract. The G4-predicting algorithm QGRS rates the ability of dysregulated lncRNAs that have been initially identified on their potential to form G4s. The subsequent clustering of lncRNAs consolidates transcript variants of each lncRNAs for the rest of this study. The functionally relevant lncRNAs within each cluster are identified using BLAST. The G4-forming capability of the lncRNAs that have been thus shortlisted is assessed and validated by a combination of CD spectroscopy, ThT fluorescence and RT stop assays. Two different in silico approaches are deployed on the G4-bearing lncRNAs to identify protein-interacting partners and shed light on their potential regulatory functions.
3. Discussion
This work is based on two primary objectives: (1) in silico identification of PQSs present in dysregulated lncRNAs of cervical cancer, and (2) in silico enunciation of G4-specific RNA-binding proteins that are likely to associate with the RNAs obtained from objective (1). The first part of our work highlights the feasibility of deploying appropriate in silico prediction methodologies for identifying G-quadruplex-forming sequences in hitherto-unexplored nucleic acid contexts. Exploration of G4 structures originated from experimental information about the behavior of specific motifs that could also be considered as reference points. The advent of multiple data repositories and structure-prediction algorithms has made it possible to develop ab initio reference points first before prioritizing experimental follow-up. At the end of our in silico pipeline, we identified 14 lncRNA clusters (
Table 1). A few lncRNAs in this list, notably MALAT1 and NEAT1, have been studied for their regulatory roles in cancer progression [
28,
29,
30]. An interesting aspect of our approach is the treatment of transcript variants of the lncRNAs selected for further scrutiny and experimental validation. It is known that there are 12 alternatively spliced variants of CRNDE, of which CRNDE-g is a highly expressed isoform in multiple cancer types [
31]. SNHG20, on the other hand, has only one variant which is upregulated in cancer and possesses a G4-forming site [
32]. Similarly, LINP1 has one predominantly expressed isoform known to adopt a stable G4 structure [
33]. In contrast, MEG3 is downregulated in cancer and has many physiologically expressed isoforms, while we are studying the variant that has PQS [
34]. Thus, the G4s being considered in the selected lncRNAs are part of functional isoforms and make our findings substantive.
We experimentally validated in silico predictions by a combination of CD spectroscopy, ThT fluorescence assay and reverse-transcriptase (RT) stop assay. As demonstrated by the in vitro experiments, the predicted putative quadruplex sequences in the four selected lncRNAs form stable G4s. Different RNA G4 topologies exhibit distinctive CD signals [
35,
36]. The orientation of strands and the molecularity of the G4s are major influences on the geometry of G4s, based on the hydrogen-bonding requirements of G-quartets and the chemical-bonding constraints of the nucleosides. CD is sensitive to the geometry of G4s and is commonly used to classify them as parallel, anti-parallel or mixed [
37]. The chosen RNA molecules adopt parallel G-quadruplexes according to their respective CD spectra. The variations in CD intensities can be attributed to varied sequence lengths, subtleties in loop lengths and overall architecture resulting in some variation in the stabilities of corresponding quadruplexes [
38,
39]. It is well known that monovalent cations can stabilize G4 structures by coordinating the O6 atom in the G-quartet channel. The inability of cations such as Li
+ to stabilize G4 formation, in contrast to the supportive role of physiologically relevant Na
+ and K
+ cations, is widely used to scrutinize the G4-forming behavior of oligonucleotides [
40]. Notably, our results suggest that while Li
+ impairs the G4-folding ability of all the selected RNAs, the presence of K
+ is most beneficial for the G4 formed by SNHG20.
Interestingly, the fluorescence enhancement of ThT was weakened in the presence of the monovalent ions for specific RNAs. While the effect of monovalent cations on DNA G4s is widely deployed as a canonical assessment of quadruplex stability, similar interpretations of RNA G4 behavior are not straight-forward. The architecture of G-tracts and spacer lengths in the MEG3, LINP1 and CRNDE-R1 sequences being tested suggest potential for polymorphism in the corresponding G4s in the presence of specific monovalent ions [
41,
42]. Considering that the CD spectra of these sequences are not significantly perturbed in the presence versus absence of K
+ or Li
+, it is possible that the parallel G4s being formed arise from a different number of participating RNA molecules. Moreover, the ThT assay relies on the dye’s ability to bind in end-stacking mode, and G4 topologies that do not provide easy access for end-stacking may be mis-identified as unstable G4s [
43,
44]. The results of ThT fluorescence assay and the RT stop assay on the selected RNAs indicates the subtle similarity in the behavior of G4s of SNHG20 and CRNDE-R2 on the one hand and MEG3, LINP1 and CRNDE-R1 on the other. The presence of two template bands in the RT-stop assay is attributable to 5′ and 3′ heterogeneity in the RNA obtained by in vitro transcription [
45,
46]. While the primary objective of our in vitro experiments was to validate the in silico searching approach, our results also point to the subtleties in in vitro behavior of the RNA G4s based on the sequence characteristics of the corresponding RNA PQSs. The value of identification and validation of G4-bearing lncRNAs in the first part of our work can be better appreciated from
Figure 8. The G4 motifs in the lncRNAs that emerge from our in silico pipeline, and that are validated through in vitro experiments, project lncRNAs, such as SNHG20, that have hitherto not been studied in the context of their secondary structure and protein interaction via such constructs in cervical cancer.
As part of our second objective, we tested two approaches to predict G4-specific RBPs that are likely to interact with the lncRNAs under study. LncRNAs are purported to exert distinctive effects via interaction with partners such as proteins, DNA, mRNA or even other lncRNAs [
47]. Among these, the identification of protein-interacting partners of a dysregulated lncRNA is likely to be of value in dissecting molecular pathways underlying cancer progression. LncRNAs have been shown to act as guides, signals, decoys and scaffolds for many proteins [
48]. RBPs are critical for regulatory RNAs to exert their cellular functions. Nevertheless, lncRNA–protein interaction can be orchestrated in many ways other than binding such as via allosteric regulatory molecules and miRNAs [
49]. Proteins such as PRC1/2, WDR5, SMAD2/3 and HnRNP are known to interact with different lncRNAs. Such lncRNA–protein associations can be connected to disease inception and propagation, thereby also providing diagnostic and therapeutic strategies for the corresponding diseases [
50,
51,
52,
53].
We employed both top-to-bottom and bottom-to-top approaches. In the top-to-bottom approach, we utilized a database called lnc2catlas, which resulted in four RBPs, TP53, CDKN2A, PTEN and SMAD4, that are ranked and categorized based on a score and their association with specific cancer types. Heatmaps were employed to analyze the co-occurrence patterns between lncRNAs and proteins. Literature mining has revealed that LINP1 does not bear the TP53-binding site to directly regulate its cellular function but p53 regulates the expression and function of LINP1 [
54]. We could not find reports confirming the direct interaction or binding of SNHG20 and CRNDE with TP53 or CDKN2A. MEG3 can interact with the p53 DNA-binding domain and its intact structure is important for p53-mediated transactivation [
55]. The negative correlation of MEG3 with CDKN2A is consistent with literature reports that suggest that the downregulation of MEG3 and overexpression of CDKN2A in cervical cancer is involved in disease progression [
56].
The inability of the top-to-bottom approach to focus exclusively on G4-binding proteins led us to test a converse bottom-to-top approach to identify the proteins that interact with RNA G4 structures. This approach relied on previously reported RNA G4-binding proteins, including FMR2, hnRNP A2, Nucleolin, DHX36, SRSF1, SRSF9, TLS and TRF2. It is intuitive to assume that the probability of binding between a lncRNA and a protein would be higher if they shared the same subcellular location. Therefore, we examined the subcellular locations of the selected lncRNAs and their interacting proteins. The in silico predictions showed colocalization between RNA-protein pairs that had attractive scores in the RPISeq analysis. Consequently, these proteins have a significant likelihood of physically interacting with lncRNAs. LINP1 is the only lncRNA having cytoplasmic presence and is known to translocate to the nucleus in response to DNA damage [
33]. It may also serve as a possible interacting partner for FMR2 and DHX36. It is worthwhile to consider that FMR2 has a nuclear localization signal and can be translocated into the nucleus or nuclear speckles if triggered by regulatory molecules [
20]. Therefore, FMR2 can also be a plausible interacting partner for CRNDE, MEG3 and SNHG20. The main takeaway from these results is the selective proteins postulated to interact with lncRNAs which can be further evaluated by in vivo proteomics experiments. The interaction of these proteins with specific lncRNAs may trigger activation or inhibition of downstream pathways that will ultimately contribute to tumor progression. The selected lncRNAs primarily participate in cell growth, epithelial-to-mesenchymal transition and apoptosis (
Table 5). Notably, among the listed RBPs in
Table 5, DHX36 has been previously reported to actively resolve G4s [
57,
58]. The other RBPs that were identified in our search are yet to be reported in direct contact with RNA G4s. Thus, these results could be used as motivation for conducting detailed experimental analyses of RBP–protein interactions.
The value of the results obtained in the second part of our work can also be better appreciated from
Figure 8. The G4 motifs in the lncRNAs that emerge from our in silico pipeline and that are validated through in vitro experiments may or may not be directly involved in associating with proteins. The presence of G4 motifs in these lncRNAs essentially serves as a “hook” to identify a host of proteins that partner with the lncRNA, and would otherwise have remained inaccessible due to the severe constraints of systematic experimental assessment. Such information is valuable for understanding the possible roles played by specific lncRNA. For example, SNHG20 is one of the four lncRNAs that we have examined for its ability to possess G4 folding sites. The identification of SNHG20 led to the subsequent prediction of interactions with TP53 and CDKN2A. Targeted experiments that probe SNHG20 interaction with TP53, CDKN2A or other proteins are likely to shed light on the biological role of SNHG20 in cervical cancer progression, which is currently not understood.
The in silico predictions in this work do not replace experimental validation. Instead, they support the in silico approach and provide a framework for systematic experimental investigations. In future, experimental validation of protein-interacting partners identified by the approach reported in this work would facilitate further scrutiny of their diagnostic and therapeutic potentials. Notably, alterations in quadruplex structure using synthetic ligands can potentially disrupt or stabilize the tertiary structure of lncRNAs, thereby affecting the lncRNA–protein partner interactions and providing a therapeutic handle. Our laboratory is currently pursuing these G4-mediated activities of dysregulated lncRNAs in cervical cancer.