Ionization Detail Parameters for DNA Damage Evaluation in Charged Particle Radiotherapy: Simulation Study Based on Cell Survival Database
Abstract
:1. Introduction
2. Results
2.1. Nanodosimetric Alternative Metric
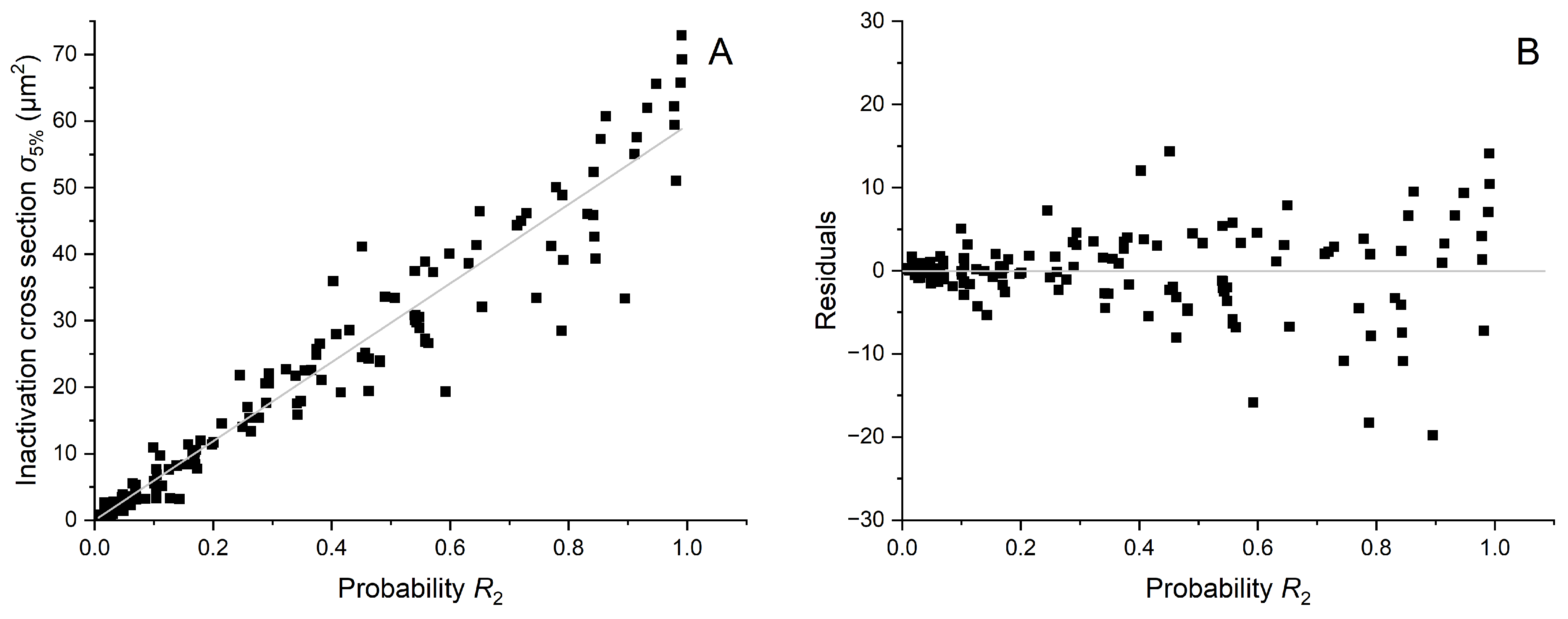
2.2. Cell Inactivation Cross Sections
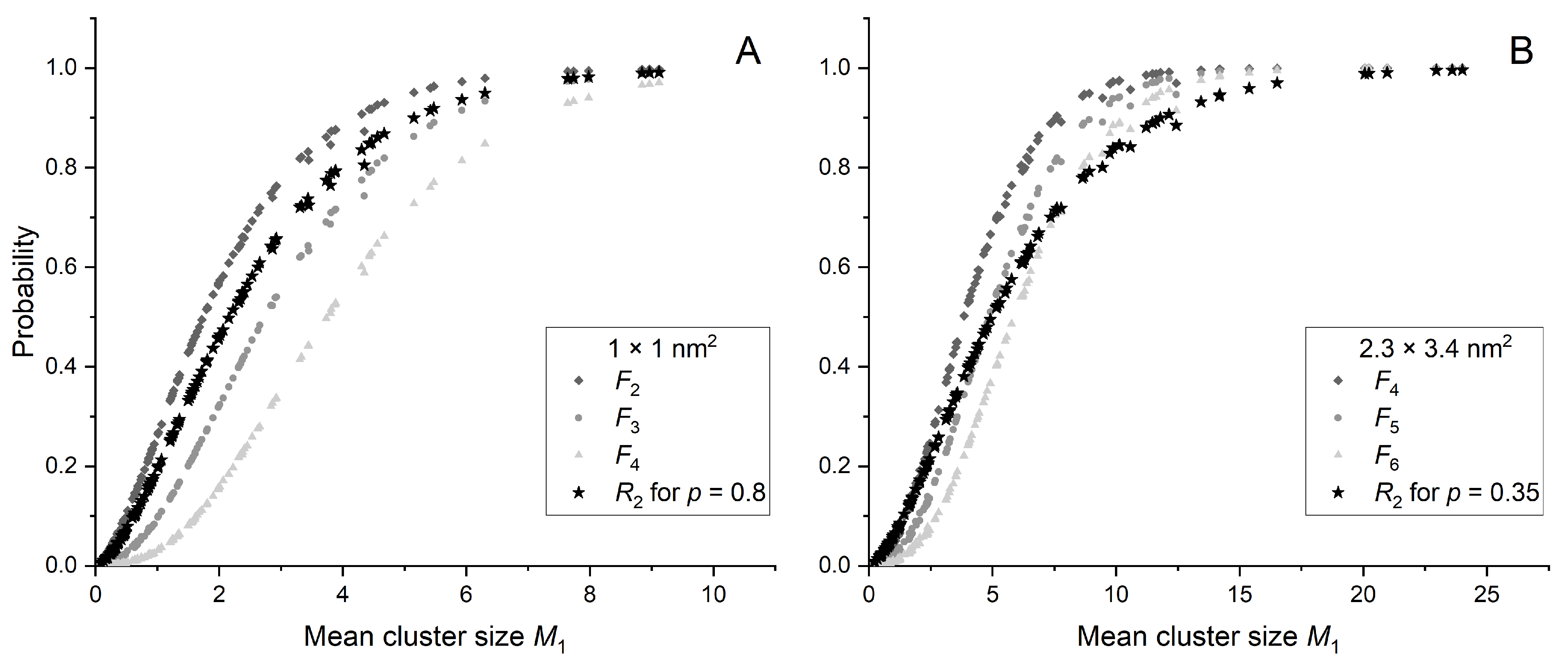
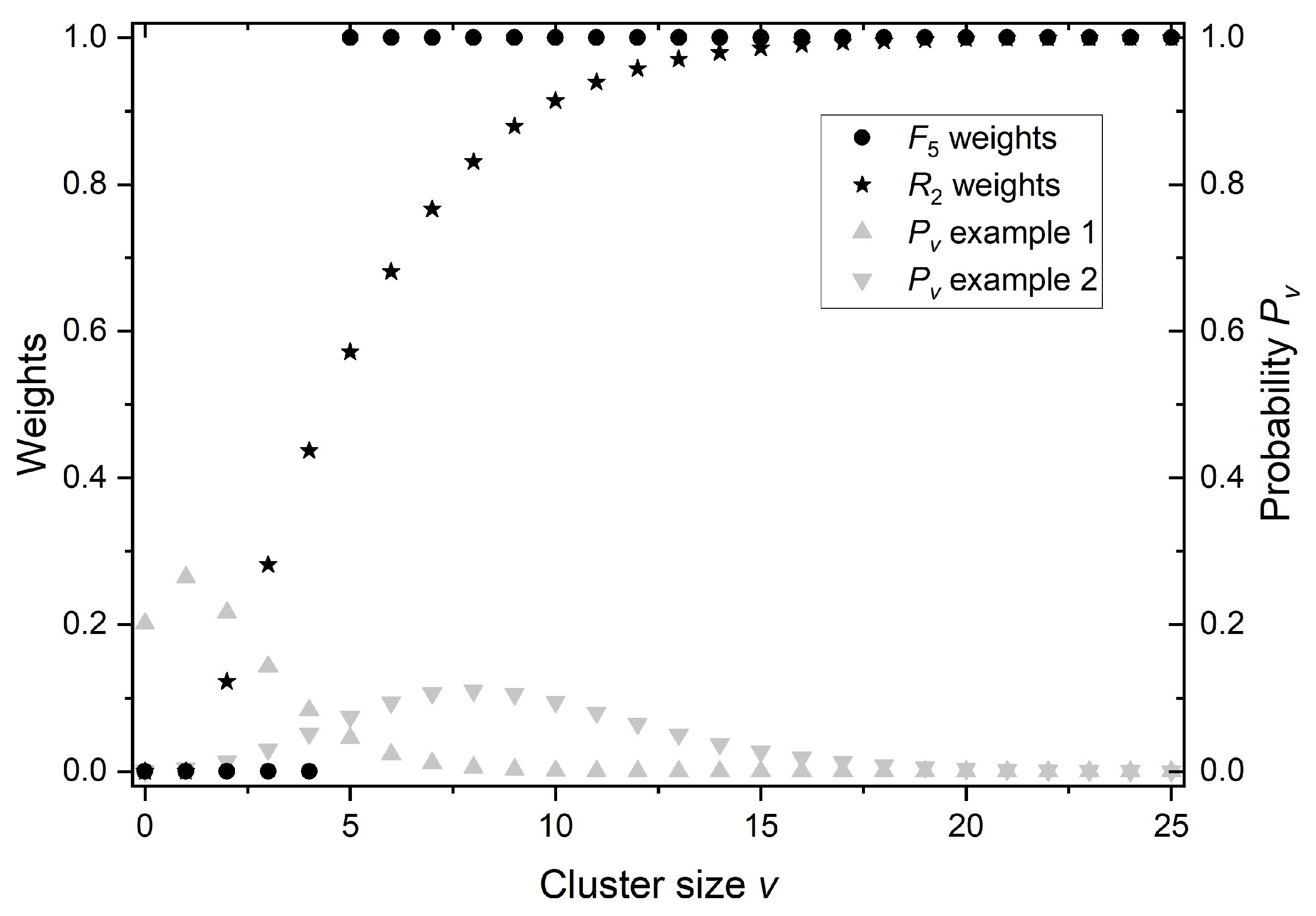
2.3. Ionization Details Parameters
2.4. Link between Radiobiology and Nanodosimetry
3. Discussion
4. Materials and Methods
4.1. Nanodosimetric Data
4.1.1. Jet Counter Nanodosimeter
4.1.2. ICSD and Other Nanodosimetric Quantities
4.1.3. Monte Carlo Simulations
4.2. Radiobiological Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DNA | Deoxyribonucleic Acid |
| DSB | Double Strand Break |
| GEM | Gas Electron Multiplier |
| GSI | GSI Helmholtz Centre for Heavy Ion Research, Darmstadt, Germany |
| ICSD | Ionization Cluster Size Distribution |
| JC | Jet Counter |
| LET | Linear energy transfer |
| LQ | Linear-Quadratic model |
| MC | Monte Carlo |
| PIDE | Particle Irradiation Data Ensemble |
| RBE | Relative Biological Effectiveness |
| SRIM | Stopping and Range of Ions in Matter |
| SSB | Single Strand Break |
Appendix A



Appendix B

References
- Goodhead, D.T. Initial Events in the Cellular Effects of Ionizing Radiations: Clustered Damage in DNA. Int. J. Radiat. Biol. 1994, 65, 7–17. [Google Scholar] [CrossRef]
- Brenner, D.; Ward, J. Constraints on energy deposition and target size of multiply damaged sites associated with DNA double-strand breaks. Int. J. Radiat. Biol. 1992, 61, 737–748. [Google Scholar] [CrossRef]
- Michalik, V. Energy Deposition Clusters in Nanometer Regions of Charged-Particle Tracks. Radiat. Res. 1993, 134, 265–270. [Google Scholar] [CrossRef]
- Schulte, R.W. Nanodosimetry: Principle and Current Status. Aip Conf. Proc. 2011, 1345, 249–261. [Google Scholar] [CrossRef]
- Palmans, H.; Rabus, H.; Belchior, A.L.; Bug, M.U.; Galer, S.; Giesen, U.; Gonon, G.; Gruel, G.; Hilgers, G.; Moro, D.; et al. Future Development of Biologically Relevant Dosimetry. Br. J. Radiol. 2015, 88, 20140392. [Google Scholar] [CrossRef]
- Conte, V.; Selva, A.; Colautti, P.; Hilgers, G.; Rabus, H.; Bantsar, A.; Pietrzak, M.; Pszona, S. Nanodosimetry: Towards a New Concept of Radiation Quality. Radiat. Prot. Dosim. 2018, 180, 150–156. [Google Scholar] [CrossRef]
- Faddegon, B.; Blakely, E.A.; Burigo, L.; Censor, Y.; Dokic, I.; Domínguez Kondo, N.; Ortiz, R.; Ramos Méndez, J.; Rucinski, A.; Schubert, K.; et al. Ionization Detail Parameters and Cluster Dose: A Mathematical Model for Selection of Nanodosimetric Quantities for Use in Treatment Planning in Charged Particle Radiotherapy. Phys. Med. Biol. 2023, 68, 175013. [Google Scholar] [CrossRef]
- Grosswendt, B. Nanodosimetry, the Metrological Tool for Connecting Radiation Physics with Radiation Biology. Radiat. Prot. Dosim. 2006, 122, 404–414. [Google Scholar] [CrossRef]
- Conte, V.; Selva, A.; Colautti, P.; Hilgers, G.; Rabus, H. Track structure characterization and its link to radiobiology. Radiat. Meas. 2017, 106, 506–511. [Google Scholar] [CrossRef]
- Rucinski, A.; Biernacka, A.; Schulte, R. Applications of nanodosimetry in particle therapy planning and beyond. Phys. Med. Biol. 2021, 66, 24TR01. [Google Scholar] [CrossRef]
- Conte, V.; Bianchi, A.; Selva, A. Track Structure of Light Ions: The Link to Radiobiology. Int. J. Mol. Sci. 2023, 24, 5826. [Google Scholar] [CrossRef] [PubMed]
- Incerti, S.; Kyriakou, I.; Bernal, M.A.; Bordage, M.C.; Francis, Z.; Guatelli, S.; Ivanchenko, V.; Karamitros, M.; Lampe, N.; Lee, S.B.; et al. Geant4-DNA Example Applications for Track Structure Simulations in Liquid Water: A Report from the Geant4-DNA Project. Med. Phys. 2018, 45, e722–e739. [Google Scholar] [CrossRef] [PubMed]
- Bernal, M.A.; Bordage, M.C.; Brown, J.M.C.; Davídková, M.; Delage, E.; Bitar, Z.E.; Enger, S.A.; Francis, Z.; Guatelli, S.; Ivanchenko, V.N.; et al. Track Structure Modeling in Liquid Water: A Review of the Geant4-DNA Very Low Energy Extension of the Geant4 Monte Carlo Simulation Toolkit. Phys. Med. Eur. J. Med. Phys. 2015, 31, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Incerti, S.; Ivanchenko, A.; Karamitros, M.; Mantero, A.; Moretto, P.; Tran, H.N.; Mascialino, B.; Champion, C.; Ivanchenko, V.N.; Bernal, M.A.; et al. Comparison of GEANT4 Very Low Energy Cross Section Models with Experimental Data in Water. Med. Phys. 2010, 37, 4692–4708. [Google Scholar] [CrossRef] [PubMed]
- Incerti, S.; Baldacchino, G.; Bernal, M.; Capra, R.; Champion, C.; Francis, Z.; Guèye, P.; Mantero, A.; Mascialino, B.; Moretto, P.; et al. The Geant4-Dna Project. Int. J. Model. Simul. Sci. Comput. 2010, 1, 157–178. [Google Scholar] [CrossRef]
- Belloni, F.; Bettega, D.; Calzolari, P.; Cherubini, R.; Massariello, P.; Tallone, L. Inactivation Cross Sections for Mammalian Cells Exposed to Charged Particles: A Phenomenological Approach. Radiat. Prot. Dosim. 2002, 99, 199–202. [Google Scholar] [CrossRef]
- Grosswendt, B. Recent Advances of Nanodosimetry. Radiat. Prot. Dosim. 2004, 110, 789–799. [Google Scholar] [CrossRef]
- Friedrich, T.; Pfuhl, T.; Scholz, M. Update of the Particle Irradiation Data Ensemble (PIDE) for Cell Survival. J. Radiat. Res. 2021, 62, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Vasi, F.; Schneider, U. First measurements of ionization cluster-size distributions with a compact nanodosimeter. Med. Phys. 2021, 48, 2566–2571. [Google Scholar] [CrossRef]
- Vasi, F.; Kempf, I.; Besserer, J.; Schneider, U. FIRE: A compact nanodosimeter detector based on ion amplification in gas. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2021, 999, 165116. [Google Scholar] [CrossRef]
- Garty, G.; Schulte, R.; Shchemelinin, S.; Grosswendt, B.; Leloup, C.; Assaf, G.; Breskin, A.; Chechik, R.; Bashkirov, V. First attempts at prediction of DNA strand-break yields using nanodosimetric data. Radiat. Prot. Dosim. 2006, 122, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Pietrzak, M.; Pszona, S.; Bantsar, A. Measurements of spatial correlations of ionisation clusters in the track of carbon ions-first results. Radiat. Prot. Dosim. 2018, 180, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Hilgers, G.; Rabus, H. Correlated ionisations in two spatially separated nanometric volumes in the track structure of 241Am alpha particles: Measurements with the PTB ion counter. Radiat. Phys. Chem. 2020, 176, 109025. [Google Scholar] [CrossRef]
- Pietrzak, M. Nanodosimetric Characteristic of Carbon Ion Beam—Experiments and Monte Carlo Simulations. Ph.D. Thesis, Faculty of Physics, University of Warsaw, Warsaw, Poland, 2023. [Google Scholar]
- Mao, H.; Zhang, H.; Luo, Y.; Yang, J.; Liu, Y.; Zhang, S.; Chen, W.; Li, Q.; Dai, Z. Primary Study of the Relative and Compound Biological Effectiveness Model for Boron Neutron Capture Therapy Based on Nanodosimetry. Med. Phys. 2024, 51, 3076–3092. [Google Scholar] [CrossRef] [PubMed]
- Durante, M. New Challenges in High-Energy Particle Radiobiology. Br. J. Radiol. 2014, 87, 20130626. [Google Scholar] [CrossRef] [PubMed]
- Mazzone, A.; Finocchiaro, P.; Meo, S.L.; Colonna, N. On the (Un)Effectiveness of Proton Boron Capture in Proton Therapy. arXiv 2018, arXiv:1802.09482. [Google Scholar] [CrossRef]
- Mastromarco, M.; Digennaro, A.; Mazzone, A.; Finocchiaro, P.; Praena, J.; Porras, I.; Colonna, N. Proton Boron Capture Therapy: Dose Calculations and a Possible New Measurement. Rad Conf. Proc. 2020, 4, 185–189. [Google Scholar] [CrossRef]
- Ahmadi Ganjeh, Z.; Mosleh-Shirazi, M.A. Macroscopic and Microscopic Investigation of Maximum Effectiveness of Proton-Boron Capture Therapy Using Monte Carlo Simulation. Radiat. Phys. Chem. 2024, 214, 111289. [Google Scholar] [CrossRef]
- Manti, L.; Attili, A.; Bláha, P.; Bortolussi, S.; Cuttone, G.; Postuma, I. Increasing Particle Therapy Biological Effectiveness by Nuclear Reaction-Driven Binary Strategies. In Monte Carlo in Heavy Charged Particle Therapy; CRC Press: Boca Raton, FL, USA, 2024. [Google Scholar]
- Safavi-Naeini, M.; Chacon, A.; Guatelli, S.; Franklin, D.R.; Bambery, K.; Gregoire, M.C.; Rosenfeld, A. Opportunistic Dose Amplification for Proton and Carbon Ion Therapy via Capture of Internally Generated Thermal Neutrons. Sci. Rep. 2018, 8, 16257. [Google Scholar] [CrossRef]
- Grosswendt, B.; De Nardo, L.; Colautti, P.; Pszona, S.; Conte, V.; Tornielli, G. Experimental Equivalent Cluster-Size Distributions in Nanometric Volumes of Liquid Water. Radiat. Prot. Dosim. 2004, 110, 851–857. [Google Scholar] [CrossRef]
- Pszona, S.; Bantsar, A.; Kula, J. Charge cluster distribution in nanosites traversed by a single ionizing particle—An experimental approach. Nucl. Instrum. Methods Phys. Res. Sect. B-Beam Interact. Mater. At. 2008, 266, 4911–4915. [Google Scholar] [CrossRef]
- Bantsar, A. Ionization Cluster Size Distributions Created by Low Energy Electrons and Alpha Particles in Nanometric Track Segment in Gases. Ph.D. Thesis, The Andrzej Sołtan Institute for Nuclear Studies, Świerk, Poland, 2010. [Google Scholar] [CrossRef]
- Bantsar, A.; Grosswendt, B.; Pszona, S.; Kula, J. Single track nanodosimetry of low energy electrons. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2009, 599, 270–274. [Google Scholar] [CrossRef]
- Bantsar, A.; Hilgers, G.; Pszona, S.; Rabus, H.; Szeflinski, Z. Experimental investigation of ionisation track structure of carbon ions at HIL Warsaw. Radiat. Prot. Dosim. 2015, 166, 253–256. [Google Scholar] [CrossRef]
- Bantsar, A.; Pietrzak, M.; Jaskóła, M.; Korman, A.; Pszona, S.; Szefliński, Z. Status report: Nanodosimetry of carbon ion beam at HIL. Rep. Pract. Oncol. Radiother. 2014, 19, S42–S46. [Google Scholar] [CrossRef]
- Selva, A.; De Nadal, V.; Cherubini, R.; Colautti, P.; Conte, V. Towards the use of nanodosimetry to predict cell survival. Radiat. Prot. Dosim. 2019, 183, 192–196. [Google Scholar] [CrossRef]
- Pietrzak, M.; Mietelska, M.; Bancer, A.; Rucinski, A.; Brzozowska, B. Geant4-DNA modeling of nanodosimetric quantities in the Jet Counter for alpha particles. Phys. Med. Biol. 2021, 66, 225008. [Google Scholar] [CrossRef]
- Agostinelli, S.; Allison, J.; Amako, K.; Apostolakis, J.; Araujo, H.; Arce, P.; Asai, M.; Axen, D.; Banerjee, S.; Barrand, G.; et al. Geant4—A simulation toolkit. Nucl. Inst. Meth. Phys. Res. A 2003, 506, 250–303. [Google Scholar] [CrossRef]
- Allison, J.; Amako, K.; Apostolakis, J.; Arce, P.; Asai, M.; Aso, T.; Bagli, E.; Bagulya, A.; Banerjee, S.; Barrand, G.; et al. Recent developments in Geant4. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2016, 835, 186–225. [Google Scholar] [CrossRef]
- Kyriakou, I.; Šefl, M.; Nourry, V.; Incerti, S. The Impact of New Geant4-DNA Cross Section Models on Electron Track Structure Simulations in Liquid Water. J. Appl. Phys. 2016, 119, 194902. [Google Scholar] [CrossRef]
- Ivanchenko, V.; Apostolakis, J.; Bagulya, A.; Abdelouahed, H.B.; Black, R.; Bogdanov, A.; Burkhard, H.; Chauvie, S.; Cirrone, P.; Cuttone, G.; et al. Recent Improvements in Geant4 Electromagnetic Physics Models and Interfaces. Prog. Nucl. Sci. Technol. 2011, 2, 898–903. [Google Scholar] [CrossRef]
- Arce, P.; Bolst, D.; Bordage, M.C.; Brown, J.M.C.; Cirrone, P.; Cortés-Giraldo, M.A.; Cutajar, D.; Cuttone, G.; Desorgher, L.; Dondero, P.; et al. Report on G4-Med, a Geant4 Benchmarking System for Medical Physics Applications Developed by the Geant4 Medical Simulation Benchmarking Group. Med. Phys. 2021, 48, 19–56. [Google Scholar] [CrossRef]
- Ziegler, J.F.; Ziegler, M.D.; Biersack, J.P. SRIM—The Stopping and Range of Ions in Matter (2010). Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 2010, 268, 1818–1823. [Google Scholar] [CrossRef]
- Friedrich, T.; Scholz, U.; ElsäSser, T.; Durante, M.; Scholz, M. Systematic Analysis of RBE and Related Quantities Using a Database of Cell Survival Experiments with Ion Beam Irradiation. J. Radiat. Res. 2013, 54, 494–514. [Google Scholar] [CrossRef]
- Hall, E.J.; Gross, W.; Dvorak, R.F.; Kellerer, A.M.; Rossi, H.H. Survival Curves and Age Response Functions for Chinese Hamster Cells Exposed to X-rays or High LET Alpha-Particles. Radiat. Res. 1972, 52, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Bird, R.P.; Burki, H.J. Survival of Synchronized Chinese Hamster Cells Exposed to Radiation of Different Linear-Energy Transfer. Int. J. Radiat. Biol. 1975, 27, 105–120. [Google Scholar] [CrossRef]
- Cox, R.; Thacker, J.; Goodhead, D.T.; Masson, W.K.; Wilkinson, R.E. Inactivation and Mutation of Cultured Mammalian Cells by Aluminium Characteristic Ultrasoft X-Rays: II. Dose-responses of Chinese Hamster and Human Diploid Cells to Aluminium X-rays and Radiations of Different LET. Int. J. Radiat. Biol. 1977, 31, 561–576. [Google Scholar] [CrossRef]
- Thacker, J.; Stretch, A.; Stephens, M.A. Mutation and Inactivation of Cultured Mammalian Cells Exposed to Beams of Accelerated Heavy Ions. II. Chinese Hamster V79 Cells. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1979, 36, 137–148. [Google Scholar] [CrossRef]
- Schlag, H.; Lücke-Huhle, C. The Influence of Ionization Density on the DNA Synthetic Phase and Survival of Irradiated Mammalian Cells. Int. J. Radiat. Biol. 1981, 40, 75–85. [Google Scholar] [CrossRef]
- Perris, A.; Pialoglou, P.; Katsanos, A.A.; Sideris, E.G. Biological Effectiveness of Low Energy Protons. I. Survival of Chinese Hamster Cells. Int. J. Radiat. Biol. 1986, 50, 1093–1101. [Google Scholar] [CrossRef]
- Folkard, M.; Prise, K.M.; Vojnovic, B.; Davies, S.; Roper, M.J.; Michael, B.D. The Irradiation of V79 Mammalian Cells by Protons with Energies below 2 Mev: Part I: Experimental Arrangement and Measurements of Cell Survival. Int. J. Radiat. Biol. 1989, 56, 221–237. [Google Scholar] [CrossRef]
- Folkard, M.; Prise, K.M.; Vojnovic, B.; Newman, H.C.; Roper, M.J.; Michael, B.D. Inactivation of V79 Cells by Low-Energy Protons, Deuterons and Helium-3 Ions. Int. J. Radiat. Biol. 1996, 69, 729–738. [Google Scholar] [CrossRef]
- Prise, K.M.; Folkard, M.; Davies, S.; Michael, B.D. The Irradiation of V79 Mammalian Cells by Protons with Energies below 2 Mev. Part II. Measurement of Oxygen Enhancement Ratios and DNA Damage. Int. J. Radiat. Biol. 1990, 58, 261–277. [Google Scholar] [CrossRef] [PubMed]
- Raju, M.R.; Eisen, Y.; Carpenter, S.; Inkret, W.C. Radiobiology of α Particles. III. Cell Inactivation by α-Particle Traversals of the Cell Nucleus. Radiat. Res. 1991, 128, 204–209. [Google Scholar] [CrossRef]
- Jenner, T.J.; DeLara, C.M.; O’Neill, P.; Stevens, D.L. Induction and Rejoining of DNA Double-Strand Breaks in V79-4 Mammalian Cells Following γ and Airradiation. Int. J. Radiat. Biol. 1993, 64, 265–273. [Google Scholar] [CrossRef]
- Weber, K.J.; Flentje, M. Lethality of Heavy Ion-Induced DNA Double-Strand Breaks in Mammalian Cells. Int. J. Radiat. Biol. 1993, 64, 169–178. [Google Scholar] [CrossRef]
- Stenerlöw, B.; Pettersson, O.A.; Essand, M.; Blomquist, E.; Carlsson, J. Irregular Variations in Radiation Sensitivity When the Linear Energy Transfer Is Increased. Radiother. Oncol. 1995, 36, 133–142. [Google Scholar] [CrossRef]
- Belli, M.; Cera, F.; Cherubini, R.; Dalla Vecchia, M.; Haqjue, A.M.; Ianzini, F.; Moschini, G.; Sapora, O.; Simone, G.; Tabocchini, M.A.; et al. RBE-LET Relationships for Cell Inactivation and Mutation Induced by Low Energy Protons in V79 Cells: Further Results at the LNL Facility. Int. J. Radiat. Biol. 1998, 74, 501–509. [Google Scholar] [CrossRef]
- Belli, M.; Bettega, D.; Calzolari, P.; Cherubini, R.; Cuttone, G.; Durante, M.; Esposito, G.; Furusawa, Y.; Gerardi, S.; Gialanella, G.; et al. Effectiveness of Monoenergetic and Spread-out Bragg Peak Carbon-Ions for Inactivation of Various Normal and Tumour Human Cell Lines. J. Radiat. Res. 2008, 49, 597–607. [Google Scholar] [CrossRef]
- Tilly, N.; Brahme, A.; Carlsson, J.; Glimelius, B. Comparison of Cell Survival Models for Mixed LET Radiation. Int. J. Radiat. Biol. 1999, 75, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Weyrather, W.K.; Ritter, S.; Scholz, M.; Kraft, G. RBE for Carbon Track-Segment Irradiation in Cell Lines of Differing Repair Capacity. Int. J. Radiat. Biol. 1999, 75, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.; Furusawa, Y.; Yamada, T. LET Dependency of Heavy-Ion Induced Apoptosis in V79 Cells. J. Radiat. Res. 2000, 41, 163–175. [Google Scholar] [CrossRef]
- Furusawa, Y.; Fukutsu, K.; Aoki, M.; Itsukaichi, H.; Eguchi-Kasai, K.; Ohara, H.; Yatagai, F.; Kanai, T.; Ando, K. Inactivation of Aerobic and Hypoxic Cells from Three Different Cell Lines by Accelerated 3He-, 12C- and 20Ne-Ion Beams. Radiat. Res. 2000, 154, 485–496. [Google Scholar] [CrossRef]
- Böhrnsen, G.; Weber, K.J.; Scholz, M. Measurement of Biological Effects of High-Energy Carbon Ions at Low Doses Using a Semi-Automated Cell Detection System. Int. J. Radiat. Biol. 2002, 78, 259–266. [Google Scholar] [CrossRef]
- Scholz, M. Effects of Ion Radiation on Cells and Tissues. In Advances in Polymer Science; Springer: Berlin/Heidelberg, Germany, 2003; Volume 162, pp. 95–155. [Google Scholar] [CrossRef]
- Hill, M.A.; Herdman, M.T.; Stevens, D.L.; Jones, N.J.; Thacker, J.; Goodhead, D.T. Relative Sensitivities of Repair-Deficient Mammalian Cells for Clonogenic Survival after α-Particle Irradiation. Radiat. Res. 2004, 162, 667–676. [Google Scholar] [CrossRef]
- Staab, A.; Zukowski, D.; Walenta, S.; Scholz, M.; Mueller-Klieser, W. Response of Chinese Hamster V79 Multicellular Spheroids Exposed to High-Energy Carbon Ions. Radiat. Res. 2004, 161, 219–227. [Google Scholar] [CrossRef]
- Zhou, G.; Kawata, T.; Furusawa, Y.; Aoki, M.; Hirayama, R.; Ando, K.; Ito, H. Protective Effects of Melatonin against Low- and High-LET Irradiation. J. Radiat. Res. 2006, 47, 175–181. [Google Scholar] [CrossRef]
- Hirayama, R.; Ito, A.; Tomita, M.; Tsukada, T.; Yatagai, F.; Noguchi, M.; Matsumoto, Y.; Kase, Y.; Ando, K.; Okayasu, R.; et al. Contributions of Direct and Indirect Actions in Cell Killing by High-LET Radiations. Radiat. Res. 2009, 171, 212–218. [Google Scholar] [CrossRef]
- Claesson, K.; Magnander, K.; Kahu, H.; Lindegren, S.; Hultborn, R.; Elmroth, K. RBE of α-Particles from 211At for Complex DNA Damage and Cell Survival in Relation to Cell Cycle Position. Int. J. Radiat. Biol. 2011, 87, 372–384. [Google Scholar] [CrossRef]
- Doria, D.; Kakolee, K.F.; Kar, S.; Litt, S.K.; Fiorini, F.; Ahmed, H.; Green, S.; Jeynes, J.C.; Kavanagh, J.; Kirby, D.; et al. Biological Effectiveness on Live Cells of Laser Driven Protons at Dose Rates Exceeding 109 Gy/s. AIP Adv. 2012, 2, 11209. [Google Scholar] [CrossRef]
- Jeynes, J.C.; Merchant, M.J.; Barazzuol, L.; Barry, M.; Guest, D.; Palitsin, V.V.; Grime, G.W.; Tullis, I.D.; Barber, P.R.; Vojnovic, B.; et al. “Broadbeam” Irradiation of Mammalian Cells Using a Vertical Microbeam Facility. Radiat. Environ. Biophys. 2013, 52, 513–521. [Google Scholar] [CrossRef]
- Tracy, B.L.; Stevens, D.L.; Goodhead, D.T.; Hill, M.A. Variation in RBE for Survival of V79-4 Cells as a Function of Alpha-Particle (Helium Ion) Energy. Radiat. Res. 2015, 184, 33–45. [Google Scholar] [CrossRef]
- Furusawa, Y.; Nakano-Aoki, M.; Matsumoto, Y.; Hirayama, R.; Kobayashi, A.; Konishi, T. Equivalency of the Quality of Sublethal Lesions after Photons and High-Linear Energy Transfer Ion Beams. J. Radiat. Res. 2017, 58, 803–808. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mietelska, M.; Pietrzak, M.; Bancer, A.; Ruciński, A.; Szefliński, Z.; Brzozowska, B. Ionization Detail Parameters for DNA Damage Evaluation in Charged Particle Radiotherapy: Simulation Study Based on Cell Survival Database. Int. J. Mol. Sci. 2024, 25, 5094. https://doi.org/10.3390/ijms25105094
Mietelska M, Pietrzak M, Bancer A, Ruciński A, Szefliński Z, Brzozowska B. Ionization Detail Parameters for DNA Damage Evaluation in Charged Particle Radiotherapy: Simulation Study Based on Cell Survival Database. International Journal of Molecular Sciences. 2024; 25(10):5094. https://doi.org/10.3390/ijms25105094
Chicago/Turabian StyleMietelska, Monika, Marcin Pietrzak, Aleksandr Bancer, Antoni Ruciński, Zygmunt Szefliński, and Beata Brzozowska. 2024. "Ionization Detail Parameters for DNA Damage Evaluation in Charged Particle Radiotherapy: Simulation Study Based on Cell Survival Database" International Journal of Molecular Sciences 25, no. 10: 5094. https://doi.org/10.3390/ijms25105094




