Natural Compounds for Bone Remodeling: A Computational and Experimental Approach Targeting Bone Metabolism-Related Proteins
Abstract
:1. Introduction
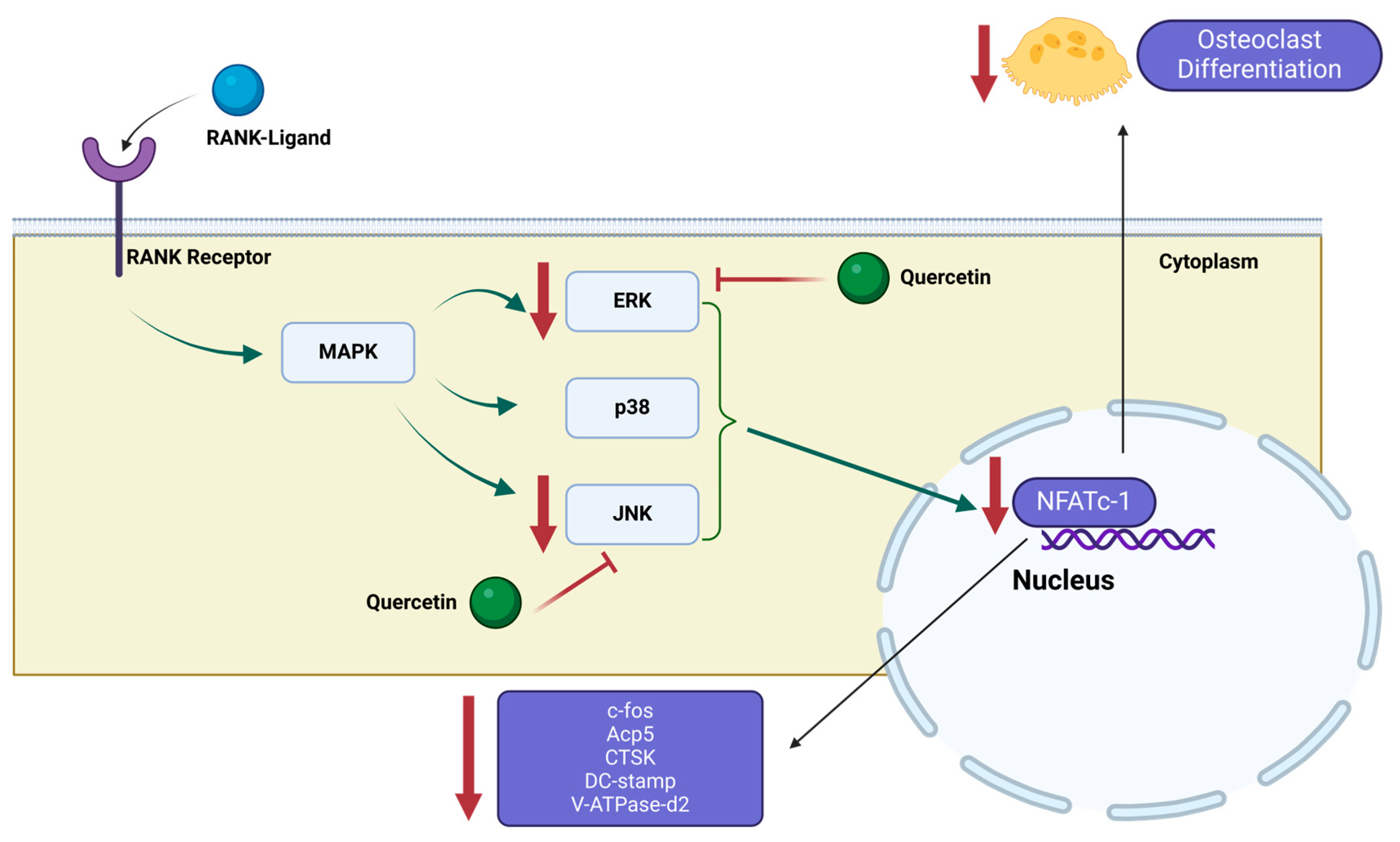
2. Results and Discussion
2.1. The Selection of the Most Promising Natural Compounds for MD Simulations
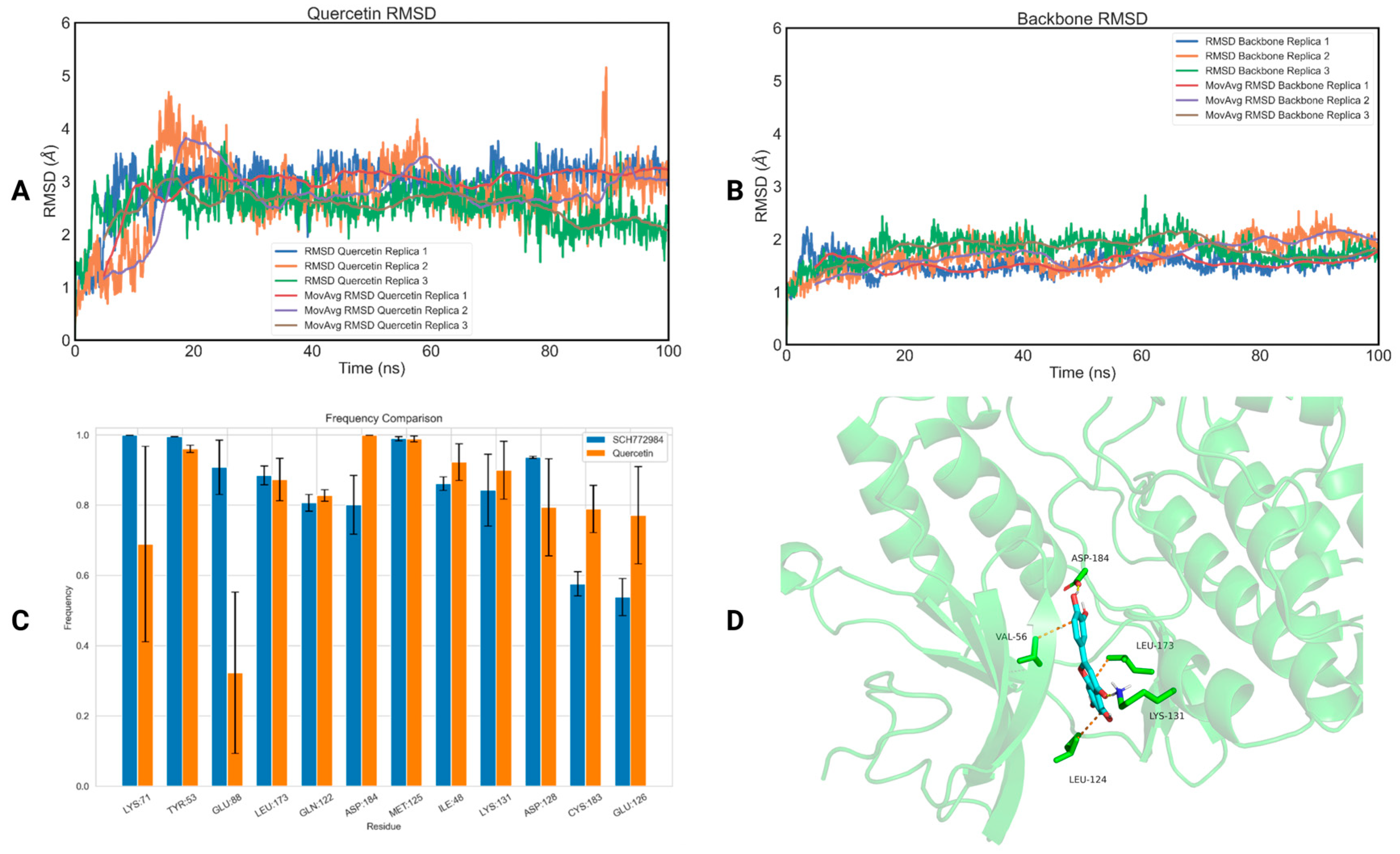
2.2. MD Simulations
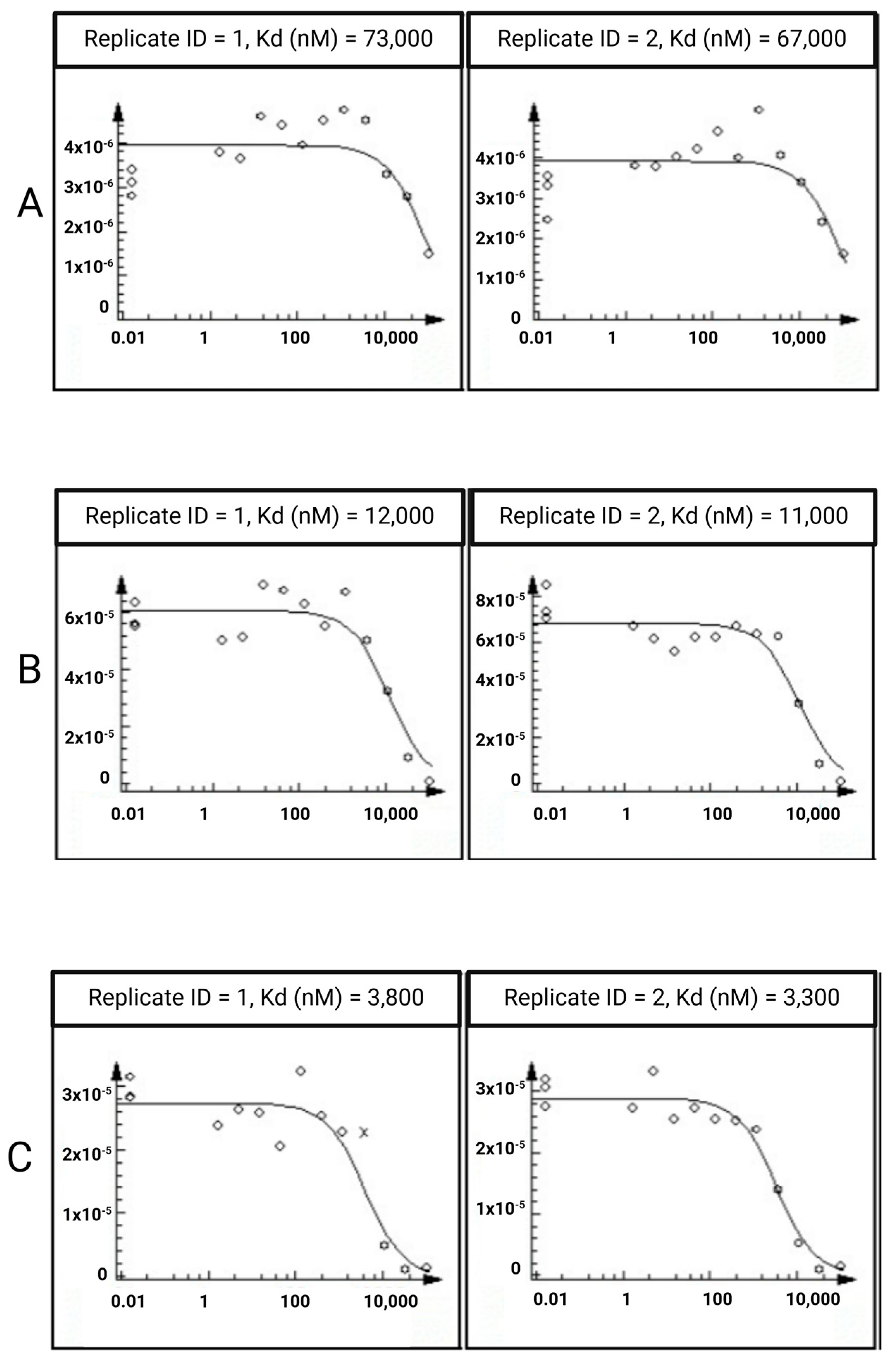
2.3. In Vitro Assays
3. Materials and Methods
3.1. The Selection of the Most Promising Natural Compounds for MD Simulations
3.2. MD Simulations
3.3. In Vitro Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- NIH Consensus Development Panel on Osteoporosis Prevention, Diagnosis, and Therapy. Osteoporosis Prevention, Diagnosis, and Therapy. JAMA 2001, 285, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Z.; Zhang, D.; Ye, D.; Zhou, Y.; Qin, J.; Zhang, Y. The prevalence and treatment rate trends of osteoporosis in postmenopausal women. PLoS ONE 2023, 18, e0290289. [Google Scholar] [CrossRef] [PubMed]
- Melton, L.J.D.; Chrischilles, E.A.; Cooper, C.; Lane, A.W.; Riggs, B.L. Perspective how many women have osteoporosis? J. Bone Miner. Res. 1992, 7, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, L.A. Secondary Causes of Osteoporosis. Mayo Clin. Proc. 2002, 77, 453–468. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Patil, S.; Jia, J. The Development of Molecular Biology of Osteoporosis. Int. J. Mol. Sci. 2021, 22, 8182. [Google Scholar] [CrossRef] [PubMed]
- Johannesdottir, F.; Thrall, E.; Muller, J.; Keaveny, T.M.; Kopperdahl, D.L.; Bouxsein, M.L. Comparison of non-invasive assessments of strength of the proximal femur. Bone 2017, 105, 93–102. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Prevention and Management of Osteoporosis; World Health Organization: Geneva, Switzerland, 2003; Volume 921, pp. 1–164. [Google Scholar]
- Kanis, J.A. Assessment of fracture risk and its application to screening for postmenopausal osteoporosis: Synopsis of a WHO report. Osteoporos. Int. 1994, 4, 368–381. [Google Scholar] [CrossRef] [PubMed]
- Kanis, J.A. Assessment of Osteoporosis at the Primary Health-Care Level. Technical Report. Available online: https://cir.nii.ac.jp/crid/1572543024192846208 (accessed on 26 January 2024).
- de Oliveira, M.A.; Moraes, R.; Castanha, E.B.; Prevedello, A.S.; Filho, J.V.; Bussolaro, F.A.; Cava, D.G. Osteoporosis Screening: Applied Methods and Technological Trends. Med. Eng. Phys. 2022, 108, 103887. [Google Scholar] [CrossRef] [PubMed]
- Sozen, T.; Ozisik, L.; Basaran, N.C. An overview and management of osteoporosis. Eur. J. Rheumatol. 2017, 4, 46–56. [Google Scholar] [CrossRef]
- Bailey, R.L.; Zou, P.; Wallace, T.C.; McCabe, G.P.; A Craig, B.; Jun, S.; A Cauley, J.; Weaver, C.M. Calcium Supplement Use Is Associated With Less Bone Mineral Density Loss, But Does Not Lessen the Risk of Bone Fracture Across the Menopause Transition: Data From the Study of Women’s Health Across the Nation. JBMR Plus 2019, 4, e10246. [Google Scholar] [CrossRef]
- Rachner, T.D.; Khosla, S.; Hofbauer, L.C. Osteoporosis: Now and the future. Lancet 2011, 377, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- De Martinis, M.; Sirufo, M.M.; Ginaldi, L. Osteoporosis: Current and Emerging Therapies Targeted to Immunological Checkpoints. Curr. Med. Chem. 2020, 27, 6356–6372. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wentworth, K.; Shoback, D.M. New Frontiers in Osteoporosis Therapy. Annu. Rev. Med. 2020, 71, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Tabatabaei-Malazy, O.; Salari, P.; Khashayar, P.; Larijani, B. New horizons in treatment of osteoporosis. DARU J. Pharm. Sci. 2017, 25, 2. [Google Scholar] [CrossRef] [PubMed]
- Mangano, G.R.A.; Avola, M.; Blatti, C.; Caldaci, A.; Sapienza, M.; Chiaramonte, R.; Vecchio, M.; Pavone, V.; Testa, G. Non-Adherence to Anti-Osteoporosis Medication: Factors Influencing and Strategies to Overcome It. A Narrative Review. J. Clin. Med. 2022, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, Z.; Li, Z.; Zhang, B.; Yao, P.; Qiao, Y. Therapeutic approach of natural products that treat osteoporosis by targeting epigenetic modulation. Front. Genet. 2023, 14, 1182363. [Google Scholar] [CrossRef] [PubMed]
- Steinhart, Z.; Angers, S. Wnt signaling in development and tissue homeostasis. Development 2018, 145, dev146589. [Google Scholar] [CrossRef] [PubMed]
- Ke, H.Z.; Richards, W.G.; Li, X.; Ominsky, M.S. Sclerostin and Dickkopf-1 as Therapeutic Targets in Bone Diseases. Endocr. Rev. 2012, 33, 747–783. [Google Scholar] [CrossRef] [PubMed]
- Baron, R.; Gori, F. Targeting WNT signaling in the treatment of osteoporosis. Curr. Opin. Pharmacol. 2018, 40, 134–141. [Google Scholar] [CrossRef]
- Li, H.; Xiao, Z.; Quarles, L.D.; Li, W. Osteoporosis: Mechanism, Molecular Target and Current Status on Drug Development. Curr. Med. Chem. 2021, 28, 1489–1507. [Google Scholar] [CrossRef]
- Cummings, S.R.; Martin, J.S.; McClung, M.R.; Siris, E.S.; Eastell, R.; Reid, I.R.; Delmas, P.; Zoog, H.B.; Austin, M.; Wang, A.; et al. Denosumab for Prevention of Fractures in Postmenopausal Women with Osteoporosis. N. Engl. J. Med. 2009, 361, 756–765. [Google Scholar] [CrossRef]
- Lee, K.; Seo, I.; Choi, M.H.; Jeong, D. Roles of Mitogen-Activated Protein Kinases in Osteoclast Biology. Int. J. Mol. Sci. 2018, 19, 3004. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Chung, Y.H.; Ahn, H.; Kim, H.; Rho, J.; Jeong, D. Selective Regulation of MAPK Signaling Mediates RANKL-dependent Osteoclast Differentiation. Int. J. Biol. Sci. 2016, 12, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Carballo, E.; Gámez, B.; Ventura, F. p38 MAPK Signaling in Osteoblast Differentiation. Front. Cell Dev. Biol. 2016, 4, 40. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Hua, F.; Ding, W. Gut Microbiome and Osteoporosis. Aging Dis. 2020, 11, 438–447. [Google Scholar] [CrossRef]
- Seely, K.D.; Kotelko, C.A.; Douglas, H.; Bealer, B.; Brooks, A.E. The Human Gut Microbiota: A Key Mediator of Osteoporosis and Osteogenesis. Int. J. Mol. Sci. 2021, 22, 9452. [Google Scholar] [CrossRef] [PubMed]
- Cresci, G.A.; Bawden, E. Gut Microbiome. Nutr. Clin. Pract. 2015, 30, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Behera, J.; Ison, J.; Tyagi, S.C.; Tyagi, N. The role of gut microbiota in bone homeostasis. Bone 2020, 135, 115317. [Google Scholar] [CrossRef] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Gao, W.; Wang, B.; Zhao, H.; Zeng, Y.; Ji, Y.; Hao, D. Diversity analysis of gut microbiota in osteoporosis and osteopenia patients. PeerJ 2017, 5, e3450. [Google Scholar] [CrossRef]
- Wen, K.; Tao, L.; Tao, Z.; Meng, Y.; Zhou, S.; Chen, J.; Yang, K.; Da, W.; Zhu, Y. Fecal and Serum Metabolomic Signatures and Microbial Community Profiling of Postmenopausal Osteoporosis Mice Model. Front. Cell. Infect. Microbiol. 2020, 10, 535310. [Google Scholar] [CrossRef]
- Li, C.; Huang, Q.; Yang, R.; Dai, Y.; Zeng, Y.; Tao, L.; Li, X.; Zeng, J.; Wang, Q. Gut microbiota composition and bone mineral loss—Epidemiologic evidence from individuals in Wuhan, China. Osteoporos. Int. 2019, 30, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Suriya, U.; Mahalapbutr, P.; Rungrotmongkol, T. Integration of In Silico Strategies for Drug Repositioning towards P38α Mitogen-Activated Protein Kinase (MAPK) at the Allosteric Site. Pharmaceutics 2022, 14, 1461. [Google Scholar] [CrossRef] [PubMed]
- Lombard, C.K.; Davis, A.L.; Inukai, T.; Maly, D.J. Allosteric Modulation of JNK Docking Site Interactions with ATP-Competitive Inhibitors. Biochemistry 2018, 57, 5897–5909. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, R.S.K.; He, P.; Modi, V.; Duong-Ly, K.C.; Ma, H.; Peterson, J.R.; Dunbrack, R.L.; Levy, R.M. Conformational Analysis of the DFG-Out Kinase Motif and Biochemical Profiling of Structurally Validated Type II Inhibitors. J. Med. Chem. 2014, 58, 466–479. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wu, H.; Wang, L.; Liu, Y.; Knapp, S.; Liu, Q.; Gray, N.S. Exploration of Type II Binding Mode: A Privileged Approach for Kinase Inhibitor Focused Drug Discovery? ACS Chem. Biol. 2014, 9, 1230–1241. [Google Scholar] [CrossRef]
- Chaikuad, A.; Tacconi, E.M.C.; Zimmer, J.; Liang, Y.; Gray, N.S.; Tarsounas, M.; Knapp, S. A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics. Nat. Chem. Biol. 2014, 10, 853–860. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion on Dietary Reference Values for folate. EFSA J. 2014, 12, 3893. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Scientific Opinion on the safety and efficacy of naringin when used as a sensory additive for all animal species. EFSA J. 2011, 9, 2416. [Google Scholar] [CrossRef]
- EFSA Panel on Food Additives and Nutrient Sources added to Food (ANS); Younes, M.; Aggett, P.; Aguilar, F.; Crebelli, R.; Dusemund, B.; Filipič, M.; Frutos, M.J.; Galtier, P.; Gott, D.; et al. Scientific opinion on the safety of green tea catechins. EFSA J. 2018, 16, e05239. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion on the substantiation of a health claim related to a combination of diosmin, troxerutin and hesperidin and maintenance of normal venous tone pursuant to Article 13(5) of Regulation (EC) No 1924/2006. EFSA J. 2014, 12, 3511. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion on the substantiation of a health claim related to alpha linolenic acid and contribution to brain and nerve tissue development pursuant to Article 14 of Regulation (EC) No 1924/2006. EFSA J. 2011, 9, 2130. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion on the substantiation of a health claim related to olive (Olea europaea L.) leaf water extract and increase in glucose tolerance pursuant to Article 13(5) of Regulation (EC) No 1924/2006. EFSA J. 2014, 12, 3655. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion on the substantiation of health claims related to rutin and improvement of endothelium-dependent vasodilation (ID 1649, 1783) and protection of DNA, proteins and lipids from oxidative damage (ID 1784) pursuant to Article 13(1) of Regula. EFSA J. 2010, 8, 1751. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion on the Tolerable Upper Intake Level of eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA) and docosapentaenoic acid (DPA). EFSA J. 2012, 10, 2815. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion Part I on the substantiation of health claims related to various food(s)/food constituent(s) not supported by pertinent human data (ID 411, 559, 1174, 1184, 1197, 1380, 1409, 1656, 1667, 1670, 1763, 1767, 1806, 1884, 1908, 1997, 2141, 2159, 2243, 2244, 2325, 2331, 2333, 2336, 2652, 2717, 2727, 2752, 2788, 2861, 2870, 2885, 2894, 3077, 3101, 3516, 3595, 3726, 4252, 4288, 4290, 4406, 4509, 4709) pursuant to Article 13(1) of Regulation (EC) No 1924/2006. EFSA J. 2011, 9, 2246. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Bampidis, V.; Azimonti, G.; Bastos, M.D.L.; Christensen, H.; Dusemund, B.; Durjava, M.F.; Kouba, M.; López-Alonso, M.; Puente, S.L.; et al. Assessment of the feed additive consisting of naringin for all animal species for the renewal of its authorisation (HealthTech Bio Actives, S.L.U. (HTBA)). EFSA J. 2022, 20, e07267. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Bampidis, V.; Azimonti, G.; Bastos, M.d.L.; Christensen, H.; Kouba, M.; Durjava, M.F.; López-Alonso, M.; Puente, S.L.; Marcon, F.; et al. Safety and efficacy of a feed additive consisting of an aqueous extract of Citrus limon (L.) Osbeck (lemon extract) for use in all animal species (Nor-Feed SAS). EFSA J. 2021, 19, e06893. [Google Scholar] [CrossRef]
- Turck, D.; Bresson, J.-L.; Burlingame, B.; Dean, T.; Fairweather-Tait, S.; Heinonen, M.; Ildico Hirsch-Ernst, K.; Mangelsdorf, I.; McArdle, H.J.; Naska, A.; et al. Safety of hydroxytyrosol as a novel food pursuant to Regulation (EC) No 258/97 EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). EFSA J. 2017, 15, 4728. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Safety of synthetic trans-resveratrol as a novel food pursuant to Regulation (EC) No 258/97. EFSA J. 2016, 14, 4368. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Bampidis, V.; Azimonti, G.; Bastos, M.D.L.; Christensen, H.; Dusemund, B.; Durjava, M.F.; Kouba, M.; López-Alonso, M.; Puente, S.L.; et al. Safety of vitamin B12 (in the form of cyanocobalamin) produced by Ensifer adhaerensCNCM-I 5541 for all animal species. EFSA J. 2020, 18, e06335. [Google Scholar] [CrossRef]
- EFSA Panel on Food Additives and Flavourings (FAF); Younes, M.; Aquilina, G.; Castle, L.; Degen, G.; Engel, K.; Fowler, P.J.; Fernandez, M.J.F.; Fürst, P.; Gundert-Remy, U.; et al. Re-evaluation of neohesperidine dihydrochalcone (E 959) as a food additive. EFSA J. 2022, 20, e07595. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Food Additives and Nutrient Sources added to Food (ANS). Statement on the safety of β-carotene use in heavy smokers. EFSA J. 2012, 10, 2953. [Google Scholar] [CrossRef]
- Bellumori, M.; Cecchi, L.; Innocenti, M.; Clodoveo, M.L.; Corbo, F.; Mulinacci, N. The EFSA Health Claim on Olive Oil Polyphenols: Acid Hydrolysis Validation and Total Hydroxytyrosol and Tyrosol Determination in Italian Virgin Olive Oils. Molecules 2019, 24, 2179. [Google Scholar] [CrossRef]
- Dodier, T.; Anderson, K.L.; Bothwell, J.; Hermann, J.; Lucas, E.A.; Smith, B.J. U.S. Montmorency Tart Cherry Juice Decreases Bone Resorption in Women Aged 65–80 Years. Nutrients 2021, 13, 544. [Google Scholar] [CrossRef]
- Ornstrup, M.J.; Harsløf, T.; Kjær, T.N.; Langdahl, B.L.; Pedersen, S.B. Resveratrol Increases Bone Mineral Density and Bone Alkaline Phosphatase in Obese Men: A Randomized Placebo-Controlled Trial. J. Clin. Endocrinol. Metab. 2014, 99, 4720–4729. [Google Scholar] [CrossRef]
- Alvarez-Sala, A.; Blanco-Morales, V.; Cilla, A.; Silvestre, R.; Hernández-Álvarez, E.; Granado-Lorencio, F.; Barberá, R.; Garcia-Llatas, G. A positive impact on the serum lipid profile and cytokines after the consumption of a plant sterol-enriched beverage with a milk fat globule membrane: A clinical study. Food Funct. 2018, 9, 5209–5219. [Google Scholar] [CrossRef]
- De Souza, M.J.; Strock, N.; Williams, N.; Lee, H.; Koltun, K.; Rogers, C.; Ferruzzi, M.; Nakatsu, C.; Weaver, C. Low Dose Daily Prunes Preserve Hip Bone Mineral Density With No Impact on Body Composition in a 12-Month Randomized Controlled Trial in Postmenopausal Women: The Prune Study. Curr. Dev. Nutr. 2022, 6, 10. [Google Scholar] [CrossRef]
- Shen, C.-L.; Chyu, M.-C.; Pence, B.C.; Yeh, J.K.; Zhang, Y.; Felton, C.K.; Doctolero, S.; Wang, J.-S. Green tea polyphenols supplementation and Tai Chi exercise for postmenopausal osteopenic women: Safety and quality of life report. BMC Complement. Altern. Med. 2010, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Wattanathorn, J.; Somboonporn, W.; Sungkamanee, S.; Thukummee, W.; Muchimapura, S. A Double-Blind Placebo-Controlled Randomized Trial Evaluating the Effect of Polyphenol-Rich Herbal Congee on Bone Turnover Markers of the Perimenopausal and Menopausal Women. Oxidative Med. Cell. Longev. 2018, 2018, 2091872. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.N.T.; Thybo, C.B.; Lykkeboe, S.; Rasmussen, L.M.; Frette, X.; Christensen, L.P.; Jeppesen, P.B. Combined bioavailable isoflavones and probiotics improve bone status and estrogen metabolism in postmenopausal osteopenic women: A randomized controlled trial. Am. J. Clin. Nutr. 2017, 106, 909–920. [Google Scholar] [CrossRef] [PubMed]
- DeBar, L.L.; Ritenbaugh, C.; Aickin, M.; Orwoll, E.; Elliot, D.; Dickerson, J.; Vuckovic, N.; Stevens, V.J.; Moe, E.; Irving, L.M. YOUTH. Arch. Pediatr. Adolesc. Med. 2006, 160, 1269–1276. [Google Scholar] [CrossRef]
- Pawlowski, J.W.; Martin, B.R.; McCabe, G.P.; McCabe, L.; Jackson, G.S.; Peacock, M.; Barnes, S.; Weaver, C.M. Impact of equol-producing capacity and soy-isoflavone profiles of supplements on bone calcium retention in postmenopausal women: A randomized crossover trial. Am. J. Clin. Nutr. 2015, 102, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, F.M.; Murray, M.J.; Lewis, R.D.; A Cramer, M.; Amato, P.; Young, R.L.; Barnes, S.; Konzelmann, K.L.; Fischer, J.G.; Ellis, K.J.; et al. Clinical outcomes of a 2-y soy isoflavone supplementation in menopausal women. Am. J. Clin. Nutr. 2011, 93, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Brink, E.; Coxam, V.; Robins, S.; Wahala, K.; Cassidy, A.; Branca, F. Long-term consumption of isoflavone-enriched foods does not affect bone mineral density, bone metabolism, or hormonal status in early postmenopausal women: A randomized, double-blind, placebo controlled study. Am. J. Clin. Nutr. 2008, 87, 761–770. [Google Scholar] [CrossRef] [PubMed]
- McCoubrey, L.E.; Elbadawi, M.; Orlu, M.; Gaisford, S.; Basit, A.W. Machine Learning Uncovers Adverse Drug Effects on Intestinal Bacteria. Pharmaceutics 2021, 13, 1026. [Google Scholar] [CrossRef]
- Ozaki, D.; Kubota, R.; Maeno, T.; Abdelhakim, M.; Hitosugi, N. Association between gut microbiota, bone metabolism, and fracture risk in postmenopausal Japanese women. Osteoporos. Int. 2020, 32, 145–156. [Google Scholar] [CrossRef]
- Yan, Q.; Cai, L.; Guo, W. New Advances in Improving Bone Health Based on Specific Gut Microbiota. Front. Cell. Infect. Microbiol. 2022, 12, 821429. [Google Scholar] [CrossRef]
- Peng, J.; Yu, X.-J.; Yu, L.-L.; Tian, F.-W.; Zhao, J.-X.; Zhang, H.; Chen, W.; Zhai, Q.-X. The influence of gut microbiome on bone health and related dietary strategies against bone dysfunctions. Food Res. Int. 2021, 144, 110331. [Google Scholar] [CrossRef] [PubMed]
- Rosener, B.; Sayin, S.; O Oluoch, P.; González, A.P.G.; Mori, H.; Walhout, A.J.; Mitchell, A. Evolved bacterial resistance against fluoropyrimidines can lower chemotherapy impact in the Caenorhabditis elegans host. eLife 2020, 9, e59831. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, J.; Zhang, S.; Li, J.; Mao, L.; Yuan, Z.; Bond, P.L.; Guo, J. Non-antibiotic pharmaceuticals promote the transmission of multidrug resistance plasmids through intra- and intergenera conjugation. ISME J. 2021, 15, 2493–2508. [Google Scholar] [CrossRef] [PubMed]
- Algavi, Y.M.; Borenstein, E. A data-driven approach for predicting the impact of drugs on the human microbiome. Nat. Commun. 2023, 14, 3614. [Google Scholar] [CrossRef] [PubMed]
- Papakyriakopoulou, P.; Velidakis, N.; Khattab, E.; Valsami, G.; Korakianitis, I.; Kadoglou, N.P. Potential Pharmaceutical Applications of Quercetin in Cardiovascular Diseases. Pharmaceuticals 2022, 15, 1019. [Google Scholar] [CrossRef] [PubMed]
- Vakali, V.; Papadourakis, M.; Georgiou, N.; Zoupanou, N.; Diamantis, D.A.; Javornik, U.; Papakyriakopoulou, P.; Plavec, J.; Valsami, G.; Tzakos, A.G.; et al. Comparative Interaction Studies of Quercetin with 2-Hydroxyl-propyl-β-cyclodextrin and 2,6-Methylated-β-cyclodextrin. Molecules 2022, 27, 5490. [Google Scholar] [CrossRef]
- Georgiou, N.; Kakava, M.G.; Routsi, E.A.; Petsas, E.; Stavridis, N.; Freris, C.; Zoupanou, N.; Moschovou, K.; Kiriakidi, S.; Mavromoustakos, T. Quercetin: A Potential Polydynamic Drug. Molecules 2023, 28, 8141. [Google Scholar] [CrossRef] [PubMed]
- Dagher, O.; Mury, P.; Thorin-Trescases, N.; Noly, P.E.; Thorin, E.; Carrier, M. Therapeutic Potential of Quercetin to Alleviate Endothelial Dysfunction in Age-Related Cardiovascular Diseases. Front. Cardiovasc. Med. 2021, 8, 658400. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Available online: https://getcontacts.github.io/ (accessed on 14 March 2024).
- Kuglstatter, A.; Ghate, M.; Tsing, S.; Villaseñor, A.G.; Shaw, D.; Barnett, J.W.; Browner, M.F. X-ray crystal structure of JNK2 complexed with the p38α inhibitor BIRB796: Insights into the rational design of DFG-out binding MAP kinase inhibitors. Bioorganic Med. Chem. Lett. 2010, 20, 5217–5220. [Google Scholar] [CrossRef]
- Feng, Y.; Park, H.; Bauer, L.; Ryu, J.C.; Yoon, S.O. Thiophene-Pyrazolourea Derivatives as Potent, Orally Bioavailable, and Isoform-Selective JNK3 Inhibitors. ACS Med. Chem. Lett. 2020, 12, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Lionta, E.; Spyrou, G.; Vassilatis, D.K.; Cournia, Z. Structure-Based Virtual Screening for Drug Discovery: Principles, Applications and Recent Advances. Curr. Top. Med. Chem. 2014, 14, 1923–1938. [Google Scholar] [CrossRef] [PubMed]
- RCSB Protein Data Bank (RCSB PDB). RCSB PDB-3GP0: Crystal Structure of Human Mitogen Activated Protein Kinase 11 (p38 beta) in Complex with Nilotinib. Available online: https://www.rcsb.org/structure/3gp0 (accessed on 10 April 2023).
- Bellon, S.; Fitzgibbon, M.J.; Fox, T.; Hsiao, H.-M.; Wilson, K.P. The structure of phosphorylated P38γ is monomeric and reveals a conserved activation-loop conformation. Structure 1999, 7, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, S.D.; Redman, A.M.; Wilson, J.W.; Deanda, F.; Shotwell, J.B.; Gerding, R.; Lei, H.; Yang, B.; Stevens, K.L.; Hassell, A.M.; et al. Optimization of 4,6-bis-anilino-1H-pyrrolo [2,3-d]pyrimidine IGF-1R tyrosine kinase inhibitors towards JNK selectivity. Bioorg. Med. Chem. Lett. 2009, 19, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, H.; Tian, Y.-S.; Kawashita, N.; Takagi, T. Mordred: A molecular descriptor calculator. J. Cheminform. 2018, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Mackerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; MacKerell, A.D. Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing. J. Chem. Inf. Model. 2012, 52, 3144–3154. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Byrd, R.H.; Lu, P.; Nocedal, J.; Zhu, C. A Limited Memory Algorithm for Bound Constrained Optimization. SIAM J. Sci. Comput. 1995, 16, 1190–1208. [Google Scholar] [CrossRef]
- Zhu, C.; Byrd, R.H.; Lu, P.; Nocedal, J. Algorithm 778: L-BFGS-B. ACM Trans. Math. Softw. 1997, 23, 550–560. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Gowers, R.J.; Linke, M.; Barnoud, J.; Reddy, T.J.E.; Melo, M.N.; Seyler, S.L.; Domanski, J.; Dotson, D.L.; Buchoux, S.; Kenney, I.M.; et al. MDAnalysis: A Python Package for The Rapid Analysis of Molecular Dynamics Simulations. In Proceedings of the 15th Python in Science Conference, Austin, TX, USA, 11–17 July 2016; Volume 98, p. 105. [Google Scholar]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; Van Gunsteren, W.F.; Mark, A.E. Peptide Folding: When Simulation Meets Experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Fabian, M.A.; Biggs, W.H., III; Treiber, D.K.; Atteridge, C.E.; Azimioara, M.D.; Benedetti, M.G.; Carter, T.A.; Ciceri, P.; Edeen, P.T.; Floyd, M.; et al. A small molecule–kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005, 23, 329–336. [Google Scholar] [CrossRef] [PubMed]





| Natural Compound | 2D Structure | Total Number of Affected Bacterial Strains | Number of Affected Strains Positively Correlated with Osteoporosis | Number of Affected Strains Negatively Correlated with Osteoporosis |
|---|---|---|---|---|
| Quercetin | 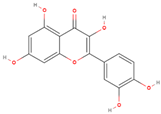 | 22 | 8 | 7 |
| Chlorogenic acid |  | 18 | 6 | 5 |
| Naringin |  | 16 | 4 | 3 |
| Hesperidin |  | 12 | 4 | 1 |
| Salidroside |  | 6 | 2 | 1 |
| Replica ID | RMSD Quercetin (Å) | RMSD Naringin (Å) | RMSD Salidroside (Å) | RMSD Chlorogenic Acid (Å) | RMSD Hesperidin (Å) |
|---|---|---|---|---|---|
| 1 | 2.96 ± 0.49 | 2.47 ± 0.57 | 8.21 ± 5.52 | 11.50 ± 1.87 | 6.15 ± 1.25 |
| 2 | 2.74 ± 0.71 | 2.49 ± 0.49 | 3.95 ± 1.29 | 5.35 ± 1.49 | 6.03 ± 1.09 |
| 3 | 2.52 ± 0.40 | 4.03 ± 1.49 | 6.33 ± 3.00 | 24.90 ± 22.93 | 6.43 ± 1.27 |
| Average RMSD | 2.74 ± 0.22 | 3.00 ± 0.89 | 6.16 ± 1.74 | 13.92 ± 10.00 | 6.20 ± 0.21 |
| Replica ID | Free Energy of Binding Quercetin (kJ/mol) | Free Energy of Binding Naringin (kJ/mol) | Free Energy of Binding SCH772984 (kJ/mol) |
|---|---|---|---|
| 1 | −16.84 ± 1.21 | +17.91 ± 5.22 | −42.65 ± 2.52 |
| 2 | −26.85 ± 2.66 | +8.45 ± 7.42 | −24.75 ± 0.93 |
| 3 | −32.94 ± 5.31 | +19.24 ± 3.53 | −34.01 ± 2.32 |
| Average free energy of binding | −25.54 ± 8.13 | +15.20 ± 5.88 | −33.80 ± 8.95 |
| Replica ID | RMSD Quercetin (Å) | RMSD Naringin (Å) | RMSD Salidroside (Å) | RMSD Chlorogenic Acid (Å) | RMSD Hesperidin (Å) |
|---|---|---|---|---|---|
| 1 | 3.40 ± 0.89 | 2.53 ± 0.53 | 3.74 ± 0.89 | 2.53 ± 0.73 | 4.06 ± 0.96 |
| 2 | 3.44 ± 0.70 | 2.51 ± 0.41 | 3.25 ± 0.70 | 1.87 ± 0.33 | 3.92 ± 0.54 |
| 3 | 2.51 ± 0.51 | 2.17 ± 0.37 | 10.31 ± 0.51 | 2.62 ± 0.47 | 3.53 ± 0.49 |
| Average RMSD | 3.12 ± 0.53 | 2.40 ± 0.20 | 5.77 ± 3.22 | 2.34 ± 0.41 | 3.84 ± 0.27 |
| Replica ID | Free Energy of Binding Quercetin (kJ/mol) | Free Energy of Binding Naringin (kJ/mol) | Free Energy of Binding Hesperidin (kJ/mol) | Free Energy of Binding Chlorogenic Acid (kJ/mol) | Free Energy of Binding Nilotinib (kJ/mol) |
|---|---|---|---|---|---|
| 1 | +2.79 ± 6.99 | −15.87 ± 5.11 | −0.57 ± 11.24 | −18.14 ± 9.92 | −3.85 ± 2.73 |
| 2 | +1.17 ± 6.11 | −18.66 ± 6.50 | −17.67 ± 9.53 | +7.15 ± 9.15 | −62.35 ± 1.54 |
| 3 | +1.80 ± 4.87 | −6.55 ± 4.14 | +3.20 ± 5.62 | +0.77 ± 5.88 | −50.62 ± 3.52 |
| Average free energy of binding | +1.92 ± 0.82 | −13.69 ± 6.34 | −5.01 ± 11.12 | −3.41 ± 13.15 | −38.94 ± 30.95 |
| Replica ID | RMSD Quercetin (Å) | RMSD Naringin (Å) |
|---|---|---|
| 1 | 2.36 ± 0.41 | 2.96 ± 0.20 |
| 2 | 1.90 ± 0.24 | 2.88 ± 0.42 |
| 3 | 1.95 ± 0.25 | 3.35 ± 0.30 |
| Average RMSD | 2.07 ± 0.25 | 3.06 ± 0.21 |
| Replica ID | Free Energy of Binding Quercetin (kJ/mol) | Free Energy of Binding Naringin (kJ/mol) | Free Energy of Binding BIRB-796 (kJ/mol) |
|---|---|---|---|
| 1 | −6.54 ± 3.44 | −16.97 ± 6.49 | −23.51 ± 2.37 |
| 2 | −7.99 ± 1.88 | −11.07 ± 8.43 | −21.65 ± 2.42 |
| 3 | +0.59 ± 2.76 | +6.17 ± 7.51 | −17.10 ± 1.79 |
| Average free energy of binding | −4.65 ± 4.59 | −7.29 ± 9.81 | −20.75 ± 2.69 |
| Replica ID | RMSD Quercetin (Å) | RMSD Naringin (Å) |
|---|---|---|
| 1 | 6.82 ± 1.44 | 3.96 ± 0.81 |
| 2 | 4.69 ± 0.85 | 5.30 ± 1.42 |
| 3 | 4.74 ± 0.56 | 8.91 ± 1.84 |
| Average RMSD | 5.41 ± 1.21 | 6.06 ± 2.09 |
| Replica ID | Free Energy of Binding Quercetin (kJ/mol) | Free Energy of Binding Naringin (kJ/mol) | Free Energy of Binding X3S (kJ/mol) |
|---|---|---|---|
| 1 | −0.52 ± 5.08 | +4.41 ± 4.58 | +5.44 ± 2.37 |
| 2 | +0.76 ± 3.51 | +9.94 ± 6.84 | −7.54 ± 5.46 |
| 3 | −0.57 ± 5.35 | +26.72 ± 7.22 | −35.79 ± 2.41 |
| Average free energy of binding | −0.11 ± 0.62 | +13.69 ± 9.49 | −12.63 ± 17.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loukas, A.-T.; Papadourakis, M.; Panagiotopoulos, V.; Zarmpala, A.; Chontzopoulou, E.; Christodoulou, S.; Katsila, T.; Zoumpoulakis, P.; Matsoukas, M.-T. Natural Compounds for Bone Remodeling: A Computational and Experimental Approach Targeting Bone Metabolism-Related Proteins. Int. J. Mol. Sci. 2024, 25, 5047. https://doi.org/10.3390/ijms25095047
Loukas A-T, Papadourakis M, Panagiotopoulos V, Zarmpala A, Chontzopoulou E, Christodoulou S, Katsila T, Zoumpoulakis P, Matsoukas M-T. Natural Compounds for Bone Remodeling: A Computational and Experimental Approach Targeting Bone Metabolism-Related Proteins. International Journal of Molecular Sciences. 2024; 25(9):5047. https://doi.org/10.3390/ijms25095047
Chicago/Turabian StyleLoukas, Alexandros-Timotheos, Michail Papadourakis, Vasilis Panagiotopoulos, Apostolia Zarmpala, Eleni Chontzopoulou, Stephanos Christodoulou, Theodora Katsila, Panagiotis Zoumpoulakis, and Minos-Timotheos Matsoukas. 2024. "Natural Compounds for Bone Remodeling: A Computational and Experimental Approach Targeting Bone Metabolism-Related Proteins" International Journal of Molecular Sciences 25, no. 9: 5047. https://doi.org/10.3390/ijms25095047






