Silicon Microring Resonator Biosensor for Detection of Nucleocapsid Protein of SARS-CoV-2
Abstract
:1. Introduction
2. Device Structure and Operating Principle
3. Fabrication
3.1. Fabrication of Si MRR
3.2. Fabrication of Flow Channel
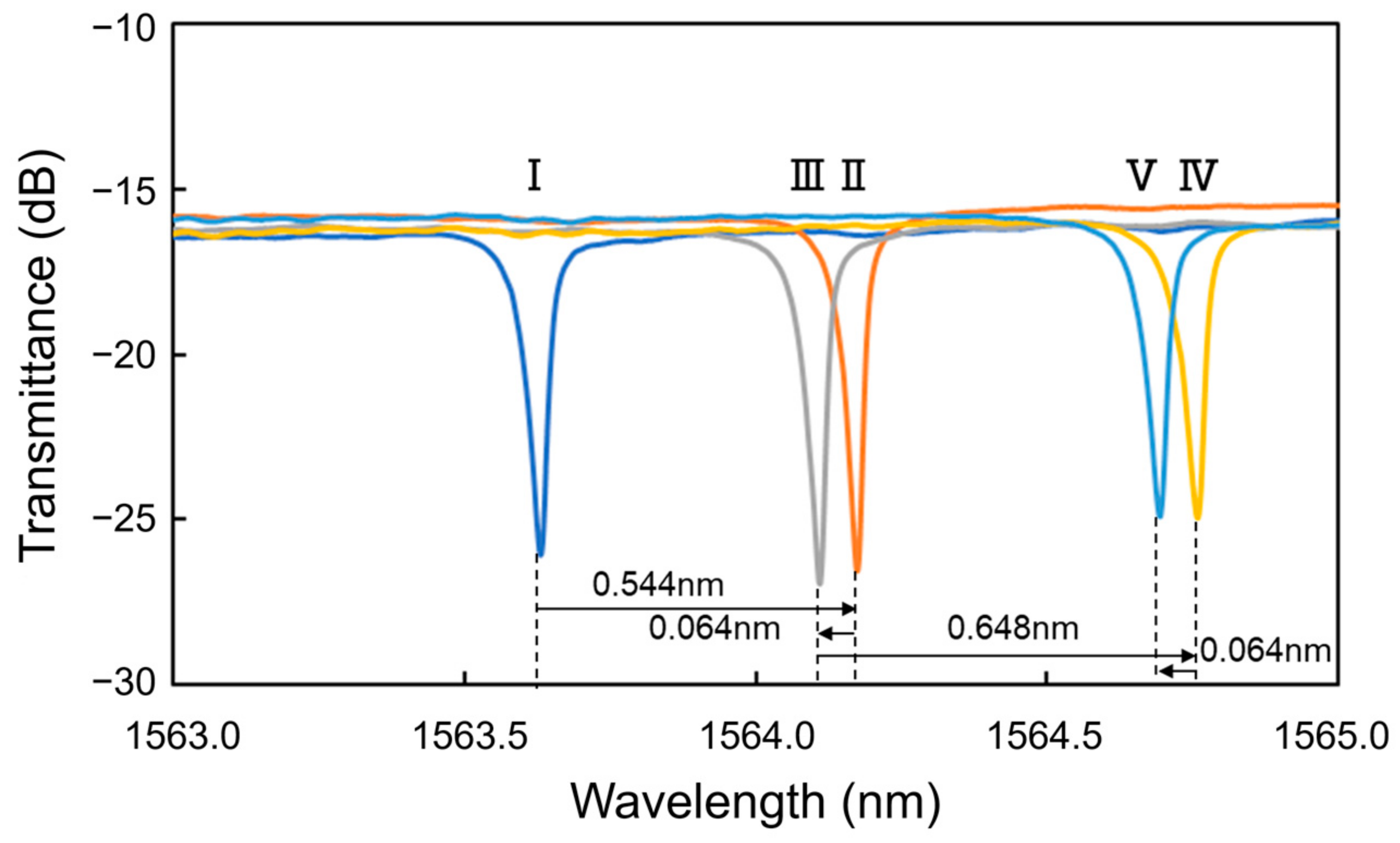
- I.
- Pouring PDMS into a flow channel mold;
- II.
- Removing air bubbles in PDMS with a vacuum pump;
- III.
- Heating in an oven at 80 °C for 2 h to harden PDMS;
- IV.
- Cutting the flow channel to the size of a single chip;
- V.
- Cleaning the chips with ammonia hydrogen peroxide mixture (APM);
- VI.
- Wiping the adhesive surface of the flow channel with ethanol-soaked cloth;
- VII.
- Ashing the bonding surface of the chip and flow channel;
- VIII.
- Attaching the flow channel to the chip;
- IX.
- Heating the chip with the flow channel in an oven at 80 °C for 20 min.
4. Detection Experiments
4.1. Modification with Antibody
- I.
- Phosphate-buffered saline (PBS) solution (pH 7.2);
- II.
- Si-taged protein G solution (0.1 mg/mL);
- II.
- PBS solution;
- IV.
- IgG antibody solution (0.01 mg/mL);
- V.
- PBS solution.
4.2. Experiments for Comparison between Specific and Nonspecific Reactions
4.3. Nucleocapsid Protein Detection Experiment
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Erdem, Ö.; Derin, E.; Sagdic, K.; Yilmaz, E.G.; Inci, F. Smart materials-integrated sensor technologies for COVID-19 diagnosis. Emergent Mater. 2021, 4, 169–185. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Z.; Zhai, X.; Li, Y.; Lin, L.; Zhao, H.; Bian, L.; Li, P.; Yu, L.; Wu, Y.; et al. Rapid and Sensitive Detection of anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal. Chem. 2020, 92, 7226–7231. [Google Scholar] [CrossRef]
- Li, R.; Tian, J.; Yang, F.; Lv, L.; Yu, J.; Sun, G.; Ma, Y.; Yang, X.; Ding, J. Clinical characteristics of 225 patients with COVID-19 in a tertiary Hospital near Wuhan, China. J. Clin. Virol. 2020, 127, 104363. [Google Scholar] [CrossRef]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS- CoV-2 and COVID-19. Nat. Rev. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Jacobs, J.L.; Haidar, G.; Mellors, J.W. COVID-19: Challenges of Viral Variants. Annu. Rev. Med. 2023, 74, 31–53. [Google Scholar] [CrossRef]
- Huang, L.; Ding, L.; Zhou, J.; Chen, S.; Chen, F.; Zhao, C.; Xu, J.; Hu, W.; Ji, J.; Xu, H.; et al. One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens. Bioelectron. 2021, 171, 112685. [Google Scholar] [CrossRef]
- Ong, V.; Soleimani, A.; Amirghasemi, F.; Nejad, S.K.; Abdelmonem, M.; Razaviyayn, M.; Hosseinzadeh, P.; Comai, L.; Mousavi, M.P.S. Impedimetric Sensing: An Emerging Tool for Combating the COVID-19 Pandemic. Biosensors 2023, 13, 204. [Google Scholar] [CrossRef]
- Hormozi Jangi, S.R. A Brief Overview of Nanozyme-Based Colorimetric and Fluorometric Sensors for Early Diagnosis of COVID-19. Transl. Med. 2023, 1, 76–84. [Google Scholar]
- Yasria, S.; Wiwanitkit, V. Sustainable materials and COVID-19 detection biosensor: A brief review. Sens. Int. 2022, 3, 100171. [Google Scholar] [CrossRef]
- Iravani, S. Nano- and biosensors for the detection of SARS-CoV-2: Challenges and opportunities. Mater. Adv. 2020, 1, 3092–3103. [Google Scholar] [CrossRef]
- Nag, P.; Sadani, K.; Mukherji, S. Optical Fiber Sensors for Rapid Screening of COVID-19. Trans. Indian Natl. Acad. Eng. 2022, 5, 233–236. [Google Scholar] [CrossRef]
- Maddali, H.; Miles, C.E.; Kohn, J.; O’Carroll, D.M. Optical Biosensors for Virus Detection: Prospects for SARS-CoV-2/COVID-19. ChemBioChem 2021, 22, 1176–1189. [Google Scholar] [CrossRef]
- Ruiz-Vega, G.; Soler, M.; Lechuga, L.M. Nanophotonic biosensors for point-of-care COVID-19 diagnostics and coronavirus surveillance. J. Phys. Photonics 2021, 3, 011002. [Google Scholar] [CrossRef]
- Vogelbacher, F.; Kothe, T.; Muellner, P.; EMelnik, E.; Sagmeister, M.; Kraft, J.; Hainberger, R. Waveguide Mach-Zehnder biosensor with laser diode pumped integrated single-mode silicon nitride organic hybrid solid-state laser. Biosens. Bioelectron. 2022, 197, 113816. [Google Scholar] [CrossRef]
- Samavati, A.; Samavati, Z.; Velashjerdi, M.; Ismail, A.F.; Othman, M.H.D.; Eisaabadi, B.G.; Abdullah, M.S.; Bolurian, M.; Bolurian, M. Sustainable and fast saliva-based COVID-19 virus diagnosis kit using a novel GO-decorated Au/FBG sensor. Chem. Eng. J. 2021, 420, 127655. [Google Scholar] [CrossRef]
- Soler, M.; Estevez, M.C.; Rubio, M.C.; Astua, A.; Lechuga, L.M. How Nanophotonic Label-Free Biosensors Can Contribute to Rapid and Massive Diagnostics of Respiratory Virus Infections: COVID-19 Case. ACS Sens. 2020, 5, 2663–2678. [Google Scholar] [CrossRef]
- Cognetti, J.S.; Steiner, D.J.; Abedin, M.; Bryan, M.R.; Shanahan, C.; Tokranova, N.; Young, E.; Klose, A.M.; Zavriyev, A.; Judy, N.; et al. Disposable photonics for cost-effective clinical bioassays: Application to COVID-19 antibody testing. Lab Chip 2021, 21, 2913–2921. [Google Scholar] [CrossRef]
- De Araújo, M.M.; da Silva, J.P. The Design of a Glycerol Concentration Sensor Based on an LRSPP Hybrid Photonic Biosensor. Sensors 2023, 23, 2010. [Google Scholar] [CrossRef]
- McClellana, M.S.; Domierb, L.L.; Baileya, R.C. Label-free virus detection using silicon photonic microring resonators. Biosens. Bioelectron. 2012, 31, 388–392. [Google Scholar] [CrossRef]
- Occhicone, A.; Sinibaldi, A.; Chiappetta, D.; Di Matteo, P.; Pileri, T.; Danz, N.; Sonntag, F.; Munzert, P.; Allegretti, M.; De Pascale, V.; et al. Detection of anti-SARS CoV-2 antibodies in human serum by means of Bloch surface waves on 1D photonic crystal biochips. Biosens. Bioelectron. X 2023, 15, 100413. [Google Scholar] [CrossRef]
- Steglich, P.; Hülsemann, M.; Dietzel, B.; Mai, A. Optical Biosensors Based on Silicon-on-Insulator Ring Resonators: A Review. Molecules 2019, 24, 519. [Google Scholar] [CrossRef]
- Matsuura, S.; Yamasaku, N.; Nishijima, Y.; Okazaki, S.; Arakawa, T. Characteristics of Highly-Sensitive Hydrogen Gas 2 Sensor Based on Pt-WO3/Si Microring Resonator. Sensors 2020, 20, 96. [Google Scholar] [CrossRef]
- Kim, G.; Son, G.; Lee, H.; Kim, K.; Lee, S. Integrated photonic glucose biosensor using a vertically coupled microring resonator in polymers. Opt. Commun. 2008, 281, 4644–4647. [Google Scholar] [CrossRef]
- Ishizaka, Y.; Makino, S.; Fujisawa, T.; Saitoh, K. A Metal-Assisted Silicon Slot Waveguide for Highly Sensitive Gas Detection. IEEE Photonics J. 2017, 9, 6800609. [Google Scholar] [CrossRef]
- Asai, N.; Sakanashi, D.; Ohashi, W.; Nakamura, A.; Kawamoto, Y.; Miyazaki, N.; Ohno, T.; Yamada, A.; Chida, S.; Shibata, Y.; et al. Efficacy and validity of automated quantitative chemiluminescent enzyme immunoassay for SARS-CoV-2 antigen test from saliva specimen in the diagnosis of COVID-19. J. Infect. Chemother. 2021, 27, 1039–1042. [Google Scholar] [CrossRef]
- Park, M.K.; Kee, J.S.; Quah, J.Y.; Netto, V.; Song, J.; Fang, Q.; Fosse, E.M.L.; Lo, G.-Q. Label-free aptamer sensor based on silicon microring resonators. Sens. Actuators B 2013, 176, 552–559. [Google Scholar] [CrossRef]
- Ciminelli, C.; Dell’Olio, F.; Conteduca, D.; Armenise, M.N. Silicon photonic biosensors. IET Optoelectron. 2019, 13, 48–54. [Google Scholar] [CrossRef]
- Ali, L.; Khan, M.; Yousuf, A.H.B.; Masud, H.; Chaudhry, M.H. IEEE Nanotechnology Symposium (ANTS) 2018. Available online: https://ieeexplore.ieee.org/xpl/conhome/8648891/proceeding (accessed on 26 September 2023).
- Dubrovsky, M.; Blevins, M.; Boriskina Svetlana, V.; Vermeulen, D. High contrast cleavage detection. Opt. Lett. 2021, 46, 2593–2596. [Google Scholar] [CrossRef]
- Uchida, Y.; Higo, A.; Arakawa, T.; Ishizaka, Y. 28th Microoptics Conference, Miyazaki, Japan, 24–27 September 2023, F-3. Available online: https://moc2023.com/ (accessed on 26 September 2023).
- Ikeda, T.; Hata, Y.; Ninomiya, K.; Ikura, Y.; Takeguchi, K.; Aoyagi, S.; Hirota, R.; Kuroda, A. Oriented immobilization of antibodies on a silicon wafer using Si-tagged protein A. Anal. Biochem. 2009, 385, 132–137. [Google Scholar] [CrossRef]
- Taniguchia, T.; Hirowataria, A.; Ikedaa, T.; Fukuyama, M.; Amemiya, Y.; Kuroda, A.; Yokoyama, S. Detection of antibody-antigen reaction by silicon nitride slot-ring biosensors using protein G. Opt. Commun. 2016, 365, 16–23. [Google Scholar] [CrossRef]
- Ikeda, T.; Kuroda, A. Why does the silica-binding protein “Si-tag” bind strongly to silica surfaces? Implications of conformational adaptation of the intrinsically disordered polypeptide to solid surfaces. Colloids Surf. B Biointerfaces 2011, 86, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Caroselli, R.; Castelló, J.G.; Escorihuela, J.; Bañuls, M.J.; Maquieira, Á.; García-Rupérez, J. Experimental Study of the Oriented Immobilization of Antibodies on Photonic Sensing Structures by Using Protein A as an Intermediate Layer. Sensors 2018, 18, 1012. [Google Scholar] [CrossRef]
- Ikeda, T.; Ueda, T.; Tajima, H.; Sekiguchi, K.; Kuroda, A. Automated enzyme-linked immunosorbent assay using beads in a single tip (BIST) technology coupled with a novel anchor protein for oriented antibody immobilization. Anal. Methods 2014, 6, 6232–6235. [Google Scholar] [CrossRef]
- Erickson, H.P. Size and Shape of Protein Molecules at the Nanometer Level Determined by Sedimentation, Gel Filtration, and Electron Microscopy. Biol. Proced. Online 2009, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Li, H.H. Refractive index of silicon and germanium and its wavelength and temperature derivatives. J. Phys. Chem. Ref. Data 1980, 9, 561–658. [Google Scholar] [CrossRef]
- Malitson, I.H. Interspecimen comparison of the refractive index of fused silica. J. Opt. Soc. Am. 1965, 55, 1205–1208. [Google Scholar] [CrossRef]
- Zhao, H.; Brown, P.H.; Schuck, P. On the Distribution of Protein Refractive Index Increments. Biophys. J. 2011, 100, 2309–2317. [Google Scholar] [CrossRef]
- Krivosudský, O.; Dráber, P.; Cifra, M. Resolving controversy of unusually high refractive index of a tubulin. Europhys. Lett. 2017, 117, 38003. [Google Scholar] [CrossRef]
- Pang, Y.; Song, H.; Cheng, W. Using optical trap to measure the refractive index of a single animal virus in culture fluid with high precision. Biomed. Opt. Express 2016, 7, 1672–1689. [Google Scholar] [CrossRef]
- Available online: https://www.mizuho-m.co.jp/en/product/files/QC_SARS-CoV-2_PI_V1.pdf (accessed on 1 March 2024).












Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uchida, Y.; Arakawa, T.; Higo, A.; Ishizaka, Y. Silicon Microring Resonator Biosensor for Detection of Nucleocapsid Protein of SARS-CoV-2. Sensors 2024, 24, 3250. https://doi.org/10.3390/s24103250
Uchida Y, Arakawa T, Higo A, Ishizaka Y. Silicon Microring Resonator Biosensor for Detection of Nucleocapsid Protein of SARS-CoV-2. Sensors. 2024; 24(10):3250. https://doi.org/10.3390/s24103250
Chicago/Turabian StyleUchida, Yusuke, Taro Arakawa, Akio Higo, and Yuhei Ishizaka. 2024. "Silicon Microring Resonator Biosensor for Detection of Nucleocapsid Protein of SARS-CoV-2" Sensors 24, no. 10: 3250. https://doi.org/10.3390/s24103250





