An Accurate Representation of the Number of bZIP Transcription Factors in the Triticum aestivum (Wheat) Genome and the Regulation of Functional Genes during Salt Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of bZIP Sequences
2.2. Transcription Factor Binding Site Prediction
2.3. Functional Gene Extraction
2.4. Phylogenetic Analysis
2.5. Principal Component Analysis of bZIP Sequences
2.6. bZIP Protein Structure Analysis
2.7. Plant Material, Growth Conditions, and Treatments
2.8. Quantitative Real-Time PCR (qPCR)
3. Results
3.1. Identification of bZIP Transcription Factors in the Wheat Genome
3.2. Domain Architecture of bZIP Transcription Factors
3.3. Phylogenetic Analysis of bZIP Transcription Factors
3.4. Principal Component Analysis
3.5. Structural Analysis of bZIP Proteins
3.6. DNA Binding Site Analysis
3.7. bZIP Family Gene Expression under Salt Stress
3.7.1. ABF2 Family
3.7.2. ABF4 Family
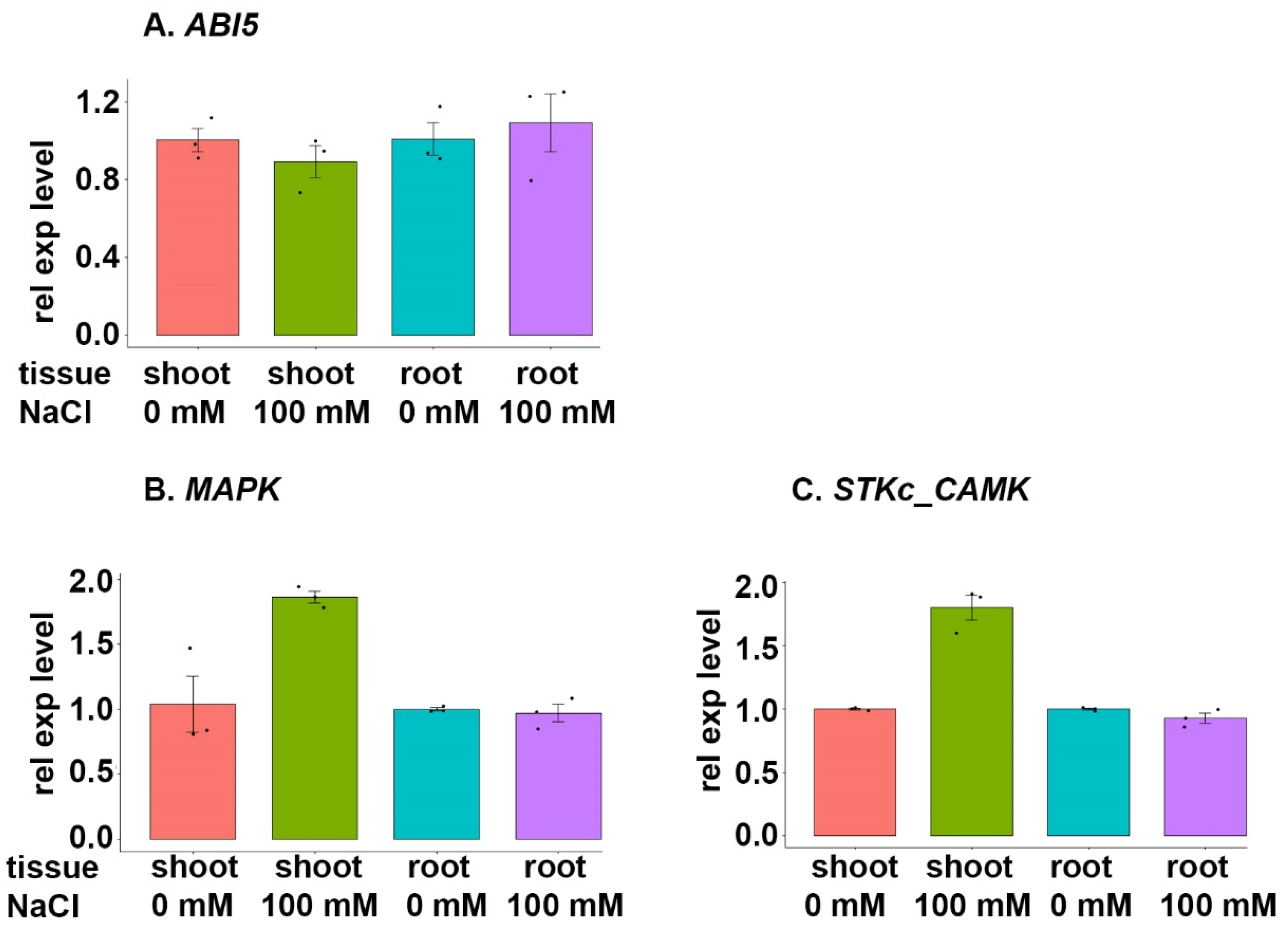
3.7.3. ABI5 Family
3.7.4. EMBP1 Family
3.7.5. VIP1 Family
4. Discussion
4.1. bZIP Expression under Salt Stress
4.2. ABF2 Family
4.3. ABF4 Family
4.4. ABI5 Family
4.5. EMPB1 Family
4.6. VIP1 Family
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Erenstein, O.; Jaleta, M.; Mottaleb, K.A.; Sonder, K.; Donovan, J.; Braun, H.-J. Global trends in wheat production, consumption and trade. In Wheat Improvement: Food Security in a Changing Climate; Springer International Publishing: Cham, Switzerland, 2022; pp. 47–66. [Google Scholar]
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef] [PubMed]
- Consortium, I.W.G.S.; Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Shankar, V.; Kumar, D.; Agrawal, V. Assessment of antioxidant enzyme activity and mineral nutrients in response to NaCl stress and its amelioration through glutathione in chickpea. Appl. Biochem. Biotechnol. 2016, 178, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Schachtman, D.; Condon, A. The significance of a two-phase growth response to salinity in wheat and barley. Funct. Plant Biol. 1995, 22, 561–569. [Google Scholar] [CrossRef]
- Liang, W.; Ma, X.; Wan, P.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Verslues, P.E.; Bailey-Serres, J.; Brodersen, C.; Buckley, T.N.; Conti, L.; Christmann, A.; Dinneny, J.R.; Grill, E.; Hayes, S.; Heckman, R.W. Burning questions for a warming and changing world: 15 unknowns in plant abiotic stress. Plant Cell 2023, 35, 67–108. [Google Scholar] [CrossRef]
- Eckardt, N.A.; Ainsworth, E.A.; Bahuguna, R.N.; Broadley, M.R.; Busch, W.; Carpita, N.C.; Castrillo, G.; Chory, J.; DeHaan, L.R.; Duarte, C.M. Climate change challenges, plant science solutions. Plant Cell 2023, 35, 24–66. [Google Scholar] [CrossRef]
- Després, C.; DeLong, C.; Glaze, S.; Liu, E.; Fobert, P.R. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 2000, 12, 279–290. [Google Scholar] [CrossRef]
- Hobo, T.; Kowyama, Y.; Hattori, T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 1999, 96, 15348–15353. [Google Scholar] [CrossRef] [PubMed]
- Oyama, T.; Shimura, Y.; Okada, K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997, 11, 2983–2995. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.J.; Burr, F.A.; Aukerman, M.J.; Burr, B. Maize regulatory gene opaque-2 encodes a protein with a" leucine-zipper" motif that binds to zein DNA. Proc. Natl. Acad. Sci. USA 1990, 87, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Sibéril, Y.; Doireau, P.; Gantet, P. Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur. J. Biochem. 2001, 268, 5655–5666. [Google Scholar] [CrossRef] [PubMed]
- Hurst, H.C. Transcription factors 1: bZIP proteins. Protein Profile 1995, 2, 101–168. [Google Scholar] [PubMed]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chu, Z. Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genom. 2015, 16, 227. [Google Scholar] [CrossRef]
- Sornaraj, P.; Luang, S.; Lopato, S.; Hrmova, M. Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: A molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Et Biophys. Acta 2016, 1860, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Ehlert, A.; Weltmeier, F.; Wang, X.; Mayer, C.S.; Smeekens, S.; Vicente-Carbajosa, J.; Dröge-Laser, W. Two-hybrid protein–protein interaction analysis in Arabidopsis protoplasts: Establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J. 2006, 46, 890–900. [Google Scholar] [CrossRef]
- Shen, H.; Cao, K.; Wang, X. AtbZIP16 and AtbZIP68, two new members of GBFs, can interact with other G group bZIPs in Arabidopsis thaliana. BMB Rep. 2008, 41, 132–138. [Google Scholar] [CrossRef]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2014, 84, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Deppmann, C.D.; Acharya, A.; Rishi, V.; Wobbes, B.; Smeekens, S.; Taparowsky, E.J.; Vinson, C. Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: A comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 2004, 32, 3435–3445. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Zou, H.-F.; Wei, W.; Hao, Y.-J.; Tian, A.-G.; Huang, J.; Liu, Y.-F.; Zhang, J.-S.; Chen, S.-Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, J.; Zhang, B.; Vanitha, J.; Ramachandran, S.; Jiang, S.Y. Genome-wide Expansion and Expression Divergence of the Basic Leucine Zipper Transcription Factors in Higher Plants with an Emphasis on Sorghum F. J. Integr. Plant Biol. 2011, 53, 212–231. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gao, S.; Tang, Y.; Li, L.; Zhang, F.; Feng, B.; Fang, Z.; Ma, L.; Zhao, C. Genome-wide identification and evolutionary analyses of bZIP transcription factors in wheat and its relatives and expression profiles of anther development related TabZIP genes. BMC Genom. 2015, 16, 976. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Yang, H.; Yan, Y.; Wei, Y.; Tie, W.; Ding, Z.; Zuo, J.; Peng, M.; Li, K. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 2016, 6, 22783. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, L.G.G.; Riaño-Pachón, D.M.; Schrago, C.G.; Vicentini dos Santos, R.; Mueller-Roeber, B.; Vincentz, M. The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PLoS ONE 2008, 3, e2944. [Google Scholar] [CrossRef]
- Pfeifer, M.; Kugler, K.G.; Sandve, S.R.; Zhan, B.; Rudi, H.; Hvidsten, T.R.; Consortium, I.W.G.S.; Mayer, K.F.; Olsen, O.-A. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 2014, 345, 1250091. [Google Scholar] [CrossRef]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; International Wheat Genome Sequencing Consortium; Jakobsen, K.S.; Wulff, B.B.; Steuernagel, B.; Mayer, K.F. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef]
- Choulet, F.; Alberti, A.; Theil, S.; Glover, N.; Barbe, V.; Daron, J.; Pingault, L.; Sourdille, P.; Couloux, A.; Paux, E. Structural and functional partitioning of bread wheat chromosome 3B. Science 2014, 345, 1249721. [Google Scholar] [CrossRef]
- Ling, H.-Q.; Zhao, S.; Liu, D.; Wang, J.; Sun, H.; Zhang, C.; Fan, H.; Li, D.; Dong, L.; Tao, Y. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 2013, 496, 87–90. [Google Scholar] [CrossRef]
- Luo, M.-C.; Gu, Y.Q.; You, F.M.; Deal, K.R.; Ma, Y.; Hu, Y.; Huo, N.; Wang, Y.; Wang, J.; Chen, S. A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc. Natl. Acad. Sci. USA 2013, 110, 7940–7945. [Google Scholar] [CrossRef]
- Jia, J.; Zhao, S.; Kong, X.; Li, Y.; Zhao, G.; He, W.; Appels, R.; Pfeifer, M.; Tao, Y.; Zhang, X. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 2013, 496, 91–95. [Google Scholar] [CrossRef]
- Consortium, I.W.G.S.; Mayer, K.F.; Rogers, J.; Doležel, J.; Pozniak, C.; Eversole, K.; Feuillet, C.; Gill, B.; Friebe, B.; Lukaszewski, A.J. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Osorio, D.; Rondón-Villarrea, P.; Torres, R. Peptides: A package for data mining of antimicrobial peptides. R J. 2015, 7, 4–14. [Google Scholar] [CrossRef]
- Lethin, J.; Byrt, C.; Berger, B.; Brien, C.; Jewell, N.; Roy, S.; Mousavi, H.; Sukumaran, S.; Olsson, O.; Aronsson, H. Improved salinity tolerance-associated variables observed in EMS mutagenized wheat lines. Int. J. Mol. Sci. 2022, 23, 11386. [Google Scholar] [CrossRef]
- Lethin, J.; Shakil, S.S.M.; Hassan, S.; Sirijovski, N.; Töpel, M.; Olsson, O.; Aronsson, H. Development and characterization of an EMS-mutagenized wheat population and identification of salt-tolerant wheat lines. BMC Plant Biol. 2020, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, S.; Lethin, J.; Liu, X.; Pelc, J.; Zeng, P.; Hassan, S.; Aronsson, H. Genome-wide analysis of MYB transcription factors in the wheat genome and their roles in salt stress response. Cells 2023, 12, 1431. [Google Scholar] [CrossRef]
- de la Reguera, E.; Veatch, J.; Gedan, K.; Tully, K.L. The effects of saltwater intrusion on germination success of standard and alternative crops. Environ. Exp. Bot. 2020, 180, 104254. [Google Scholar] [CrossRef]
- Agarwal, P.; Baranwal, V.K.; Khurana, P. Genome-wide analysis of bZIP transcription factors in wheat and functional characterization of a TabZIP under abiotic stress. Sci. Rep. 2019, 9, 4608. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Xia, J.; Jiang, Y.; Bao, Y.; Chen, H.; Wang, D.; Zhang, D.; Yu, J.; Cang, J. Genome-wide identification and analysis of bZIP gene family and resistance of TaABI5 (TabZIP96) under freezing stress in wheat (Triticum aestivum). Int. J. Mol. Sci. 2022, 23, 2351. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Zhou, R.; Dossa, K.; Yu, J.; Li, D.; Liu, A.; Mmadi, M.A.; Zhang, X.; You, J. Identification and characterization of the bZIP transcription factor family and its expression in response to abiotic stresses in sesame. PLoS ONE 2018, 13, e0200850. [Google Scholar] [CrossRef] [PubMed]
- Baloglu, M.C.; Eldem, V.; Hajyzadeh, M.; Unver, T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 2014, 9, e96014. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yu, D.; Yu, J.; Zhu, D.; Zhao, Q. Functional analysis of maize silk-specific ZmbZIP25 promoter. Int. J. Mol. Sci. 2018, 19, 822. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Feng, B.; Zhang, F.; Tang, Y.; Zhang, L.; Ma, L.; Zhao, C.; Gao, S. Bioinformatic analyses of subgroup-A members of the wheat bZIP transcription factor family and functional identification of TabZIP174 involved in drought stress response. Front. Plant Sci. 2016, 7, 1643. [Google Scholar] [CrossRef]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Amirbakhtiar, N.; Ismaili, A.; Ghaffari, M.-R.; Mirdar Mansuri, R.; Sanjari, S.; Shobbar, Z.-S. Transcriptome analysis of bread wheat leaves in response to salt stress. PLoS ONE 2021, 16, e0254189. [Google Scholar] [CrossRef] [PubMed]
- Amirbakhtiar, N.; Ismaili, A.; Ghaffari, M.R.; Nazarian Firouzabadi, F.; Shobbar, Z.-S. Transcriptome response of roots to salt stress in a salinity-tolerant bread wheat cultivar. PLoS ONE 2019, 14, e0213305. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Guo, H.; Xie, Y.; Zhao, L.; Gu, J.; Zhao, S.; Li, J.; Liu, L. RNAseq analysis reveals pathways and candidate genes associated with salinity tolerance in a spaceflight-induced wheat mutant. Sci. Rep. 2017, 7, 2731. [Google Scholar] [CrossRef] [PubMed]
- Borrill, P.; Harrington, S.A.; Simmonds, J.; Uauy, C. Identification of transcription factors regulating senescence in wheat through gene regulatory network modelling. Plant Physiol. 2019, 180, 1740–1755. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.Q.; Liu, Y.F.; Liu, P.; Lei, G.; He, S.J.; Ma, B.; Zhang, W.K.; Zhang, J.S.; Chen, S.Y. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 2010, 62, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shang, J.; Chen, D.; Lei, C.; Zou, Y.; Zhai, W.; Liu, G.; Xu, J.; Ling, Z.; Cao, G. AB-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006, 46, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Czernic, P.; Visser, B.; Sun, W.; Savouré, A.; Deslandes, L.; Marco, Y.; Van Montagu, M.; Verbruggen, N. Characterization of an Arabidopsis thaliana receptor-like protein kinase gene activated by oxidative stress and pathogen attack. Plant J. 1999, 18, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Meng, Q.; Xu, J.; Tang, H.; Tang, S.; Zhang, H.; Huang, J. Knock-down of stress inducible OsSRFP1 encoding an E3 ubiquitin ligase with transcriptional activation activity confers abiotic stress tolerance through enhancing antioxidant protection in rice. Plant Mol. Biol. 2015, 87, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Kumar, R.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Overexpression of rice CBS domain containing protein improves salinity, oxidative, and heavy metal tolerance in transgenic tobacco. Mol. Biotechnol. 2012, 52, 205–216. [Google Scholar] [CrossRef]
- Zhou, Y.-B.; Liu, C.; Tang, D.-Y.; Yan, L.; Wang, D.; Yang, Y.-Z.; Gui, J.-S.; Zhao, X.-Y.; Li, L.-G.; Tang, X.-D. The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 2018, 30, 1100–1118. [Google Scholar] [CrossRef]
- Shariatipour, N.; Heidari, B. Meta-analysis of expression of the stress tolerance associated genes and uncover their-regulatory elements in rice (Oryza sativa L.). Open Bioinform. J. 2020, 13, 39–49. [Google Scholar] [CrossRef]
- Aziz, A.; Sabeem, M.; Mullath, M.; Kutty, S.; Faical, B.; Khaled, M. Plant group II LEA proteins: Intrinsically disordered structure for multiple functions in response to environmental stresses. Biomolecules 2021, 11, 1662. [Google Scholar] [CrossRef]
- Meng, Y.-C.; Zhang, H.-F.; Pan, X.-X.; Chen, N.; Hu, H.-F.; Haq, S.u.; Khan, A.; Chen, R.-G. CaDHN3, a pepper (Capsicum annuum L.) dehydrin gene enhances the tolerance against salt and drought stresses by reducing ROS accumulation. Int. J. Mol. Sci. 2021, 22, 3205. [Google Scholar] [CrossRef]
- Qin, Y.-x.; Qin, F. Dehydrins from wheat x Thinopyrum ponticum amphiploid increase salinity and drought tolerance under their own inducible promoters without growth retardation. Plant Physiol. Biochem. 2016, 99, 142–149. [Google Scholar] [CrossRef]
- Gu, L.; Liu, Y.; Zong, X.; Liu, L.; Li, D.-P.; Li, D.-Q. Overexpression of maize mitogen-activated protein kinase gene, ZmSIMK1 in Arabidopsis increases tolerance to salt stress. Mol. Biol. Rep. 2010, 37, 4067–4073. [Google Scholar] [CrossRef]
- Mishra, N.S.; Tuteja, R.; Tuteja, N. Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 2006, 452, 55–68. [Google Scholar] [CrossRef]
- Pitzschke, A.; Schikora, A.; Hirt, H. MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 2009, 12, 421–426. [Google Scholar] [CrossRef]
- Xu, H.; Li, K.; Yang, F.; Shi, Q.; Wang, X. Overexpression of CsNMAPK in tobacco enhanced seed germination under salt and osmotic stresses. Mol. Biol. Rep. 2010, 37, 3157–3163. [Google Scholar] [CrossRef]
- Kumar, K.; Raina, S.K.; Sultan, S.M. Arabidopsis MAPK signaling pathways and their cross talks in abiotic stress response. J. Plant Biochem. Biotechnol. 2020, 29, 700–714. [Google Scholar] [CrossRef]
- Wang, G.; Liang, Y.-h.; Zhang, J.-y.; Cheng, Z.-M. Cloning, molecular and functional characterization by overexpression in Arabidopsis of MAPKK genes from grapevine (Vitis vinifera). BMC Plant Biol. 2020, 20, 194. [Google Scholar] [CrossRef]
- Kumar, R.R.; Arora, K.; Goswami, S.; Sakhare, A.; Singh, B.; Chinnusamy, V.; Praveen, S. MAPK enzymes: A ROS activated signaling sensors involved in modulating heat stress response, tolerance and grain stability of wheat under heat stress. 3 Biotech 2020, 10, 380. [Google Scholar] [CrossRef] [PubMed]
- Harrington, S.A.; Backhaus, A.E.; Singh, A.; Hassani-Pak, K.; Uauy, C. The wheat GENIE3 network provides biologically-relevant information in polyploid wheat. G3 Genes Genomes Genet. 2020, 10, 3675–3686. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-González, R.; Borrill, P.; Lang, D.; Harrington, S.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; Van Ex, F.; Pasha, A. The transcriptional landscape of polyploid wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef] [PubMed]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. expVIP: A customizable RNA-seq data analysis and visualization platform. Plant Physiol. 2016, 170, 2172–2186. [Google Scholar] [CrossRef] [PubMed]
- Borrill, P.; Adamski, N.; Uauy, C. Genomics as the key to unlocking the polyploid potential of wheat. New Phytol. 2015, 208, 1008–1022. [Google Scholar] [CrossRef] [PubMed]
- Ciftci-Yilmaz, S.; Mittler, R. The zinc finger network of plants. Cell. Mol. Life Sci. 2008, 65, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, L.; Pan, Z.; Zhao, M.; Zhu, L.; Han, Y.; Li, L.; Wang, Y.; Wang, K.; Liu, S. Genome-wide analysis of the C2H2 zinc finger protein gene family and its response to salt stress in ginseng, Panax ginseng Meyer. Sci. Rep. 2022, 12, 10165. [Google Scholar] [CrossRef] [PubMed]
- Kiełbowicz-Matuk, A. Involvement of plant C2H2-type zinc finger transcription factors in stress responses. Plant Sci. 2012, 185, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Pang, J.; Zhang, F.; Sun, L.; Yang, L.; Siddique, K.H. Transcriptomic and metabolomics-based analysis of key biological pathways reveals the role of lipid metabolism in response to salt stress in the root system of Brassica napus. Plant Growth Regul. 2022, 97, 127–141. [Google Scholar] [CrossRef]
- John, R.; Raja, V.; Ahmad, M.; Jan, N.; Majeed, U.; Ahmad, S.; Yaqoob, U.; Kaul, T. Trehalose: Metabolism and role in stress signaling in plants. In Stress Signaling in Plants: Genomics and Proteomics Perspective, Volume 2; Springer: Cham, Switzerland, 2017; pp. 261–275. [Google Scholar]
- Kusano, T.; Berberich, T.; Tateda, C.; Takahashi, Y. Polyamines: Essential factors for growth and survival. Planta 2008, 228, 367–381. [Google Scholar] [CrossRef]
- Ebeed, H.T.; Hassan, N.M.; Aljarani, A.M. Exogenous applications of polyamines modulate drought responses in wheat through osmolytes accumulation, increasing free polyamine levels and regulation of polyamine biosynthetic genes. Plant Physiol. Biochem. 2017, 118, 438–448. [Google Scholar] [CrossRef]
- Nahar, K.; Hasanuzzaman, M.; Rahman, A.; Alam, M.M.; Mahmud, J.-A.; Suzuki, T.; Fujita, M. Polyamines confer salt tolerance in mung bean (Vigna radiata L.) by reducing sodium uptake, improving nutrient homeostasis, antioxidant defense, and methylglyoxal detoxification systems. Front. Plant Sci. 2016, 7, 1104. [Google Scholar] [CrossRef] [PubMed]
- Golldack, D.; Lüking, I.; Yang, O. Plant tolerance to drought and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef]
- Century, K.; Reuber, T.L.; Ratcliffe, O.J. Regulating the regulators: The future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol. 2008, 147, 20–29. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Qu, S.-C.; Du, X.-L.; Qiao, Y.-S.; Cai, B.-H.; Guo, Z.-R.; Zhang, Z. Overexpression of the Malus hupehensis MhTGA2 gene, a novel bZIP transcription factor for increased tolerance to salt and osmotic stress in transgenic tobacco. Int. J. Plant Sci. 2012, 173, 441–453. [Google Scholar] [CrossRef]









| IDs | Protein Name | Organism | DNA Binding Site |
|---|---|---|---|
| 1A02 | NFAT, FOS AND JUN | Homo sapiens | ATTTGTTTC |
| 1DH3 | CREB bZIP-CRE COMPLEX | Mus musculus | TGACGTC |
| 1FOS | C-FOS:C-JUN | Homo sapiens | AGTC |
| 1GD2 | bZIP TRANSCRIPTION FACTOR PAP1 | Schizosaccharomyces pombe (strain 972/ATCC 24843) | AGGTTACGTAA |
| 1JNM | Jun/CRE Complex | Homo sapiens | TCGATGA |
| 1S9K | Human NFAT1 and Fos-Jun | Homo sapiens | ATATGTGTA |
| 1T2K | RF3, ATF-2 and Jun | Homo sapiens | AAATGAC |
| 2H7H | JUN bZIP homodimer | Avian sarcoma virus (strain 17) | CGATGAC |
| 2WT7 | MafB:cFos | Synthetic construct, Mus musculus | ATTGCTGAC |
| 5T01 | Human c-Jun | Synthetic construct, Homo sapiens | CTATGA |
| 5VPE | FosB/JunD bZIP | Homo sapiens | GGTGACTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Sukumaran, S.; Viitanen, E.; Naik, N.; Hassan, S.; Aronsson, H. An Accurate Representation of the Number of bZIP Transcription Factors in the Triticum aestivum (Wheat) Genome and the Regulation of Functional Genes during Salt Stress. Curr. Issues Mol. Biol. 2024, 46, 4417-4436. https://doi.org/10.3390/cimb46050268
Liu X, Sukumaran S, Viitanen E, Naik N, Hassan S, Aronsson H. An Accurate Representation of the Number of bZIP Transcription Factors in the Triticum aestivum (Wheat) Genome and the Regulation of Functional Genes during Salt Stress. Current Issues in Molecular Biology. 2024; 46(5):4417-4436. https://doi.org/10.3390/cimb46050268
Chicago/Turabian StyleLiu, Xin, Selvakumar Sukumaran, Esteri Viitanen, Nupur Naik, Sameer Hassan, and Henrik Aronsson. 2024. "An Accurate Representation of the Number of bZIP Transcription Factors in the Triticum aestivum (Wheat) Genome and the Regulation of Functional Genes during Salt Stress" Current Issues in Molecular Biology 46, no. 5: 4417-4436. https://doi.org/10.3390/cimb46050268





