Complete Plastid Genome Sequences of Three Tropical African Beilschmiediineae Trees (Lauraceae: Crytocaryeae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling, DNA Extraction, Genomic Sequencing
2.2. Genome Assembly and Annotation
2.3. Phylogenetic Analysis
2.4. Analysis of Repeats and Simple Sequence Repeats (SSR)
2.5. Mutation Events Detection and Sliding Window Analysis of the Plastomes
2.6. Selective Pressure Analysis
3. Results
3.1. Phylogenetic Analysis
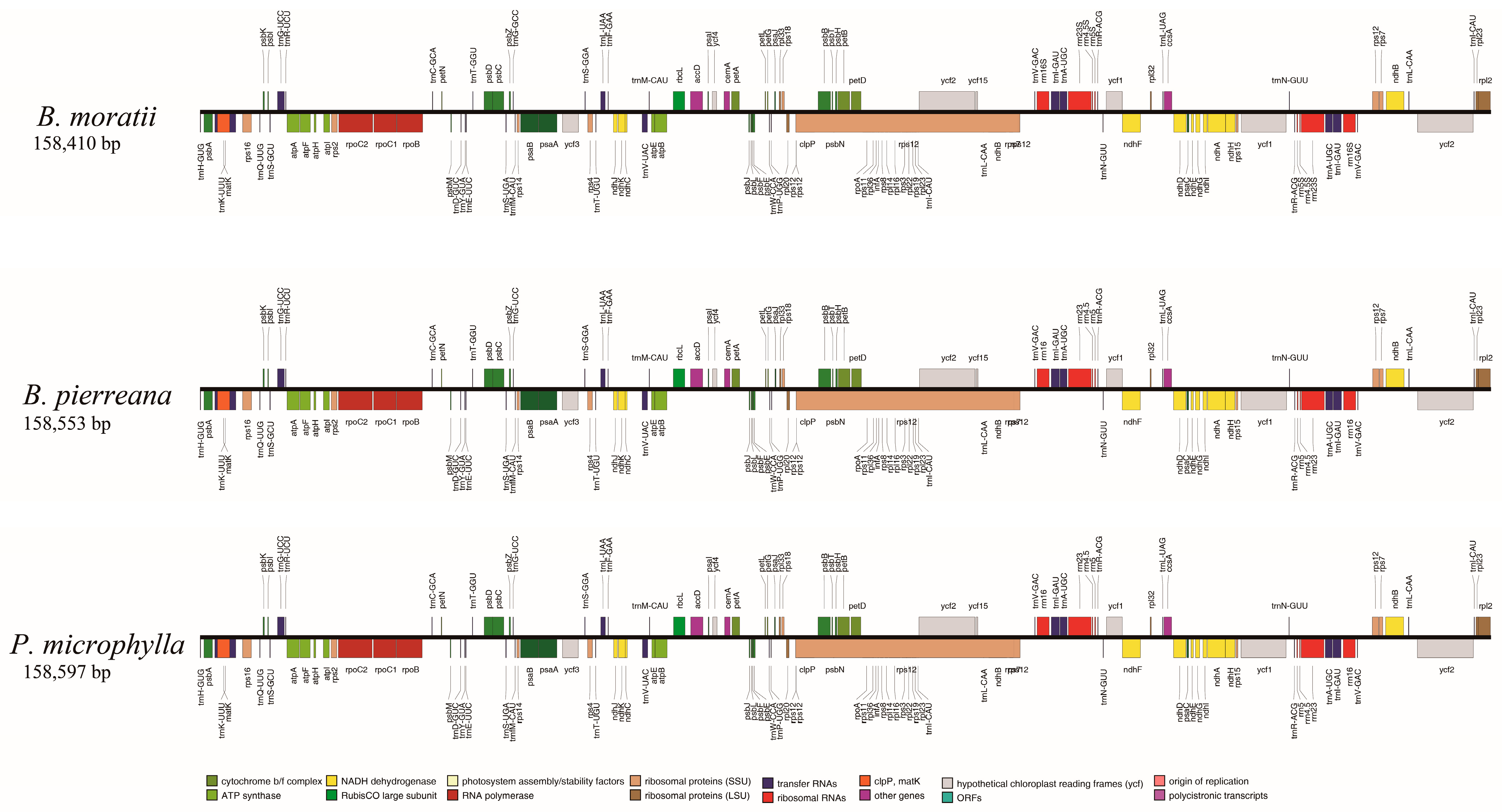
3.2. Characteristics of the cp Genome of B. moratii
3.3. Repeats Analysis
3.4. Number and Forms of Microstructural Mutations
3.5. Numbers and Pattern of SNP Mutations
3.6. Divergence Hotspots of B. moratii
3.7. Selective Pressure Analysis of Protein-Coding Genes
4. Discussion
4.1. Phylogenetic Analysis of Beilschmiediineae from Africa
4.2. Structural Variation of B. moratii Plastome
4.3. Divergence Hotspot Analysis
4.4. Selective Pressure Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Ralimanana, H.; Perrigo, A.L.; Smith, R.J.; Borrell, J.S.; Faurby, S.; Rajaonah, M.T.; Randriamboavonjy, T.; Vorontsova, M.S.; Cooke, R.S.C.; Phelps, L.N.; et al. Madagascar’s extraordinary biodiversity: Threats and opportunities. Science 2022, 378, eadf1466. [Google Scholar] [CrossRef] [PubMed]
- Marchese, C. Biodiversity hotspots: A shortcut for a more complicated concept. Glob. Ecol. Conserv. 2015, 3, 297–309. [Google Scholar] [CrossRef]
- Schettino, A.; Scotese, C.R. Apparent polar wander paths for the major continents (200 Ma to the present day): A palaeomagnetic reference frame for global plate tectonic reconstructions. Geophys. J. Int. 2005, 163, 727–759. [Google Scholar] [CrossRef]
- Ali, J.R.; Aitchison, J.C. Gondwana to Asia: Plate tectonics, paleogeography and the biological connectivity of the Indian sub-continent from the Middle Jurassic through latest Eocene (166–35 Ma). Earth-Sci. Rev. 2008, 88, 145–166. [Google Scholar] [CrossRef]
- Emanuel, K. Tropical Cyclones. Annu. Rev. Earth Planet. Sci. 2003, 31, 75–104. [Google Scholar] [CrossRef]
- Madagascar Catalogue. Catalogue of the Plants of Madagascar (Missouri Botanical Garden). Available online: https://www.tropicos.org/Project/Madagascar (accessed on 23 March 2024).
- Liu, Y.; Wang, Y.; Willett, S.D.; Zimmermann, N.E.; Pellissier, L. Escarpment evolution drives the diversification of the Madagascar flora. Science 2024, 383, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Blanco, M.B.; Greene, L.K.; Rasambainarivo, F.; Toomey, E.; Williams, R.C.; Andrianandrasana, L.; Larsen, P.A.; Yoder, A.D. Next-generation technologies applied to age-old challenges in Madagascar. Conserv. Genet. 2020, 21, 785–793. [Google Scholar] [CrossRef]
- Krehenwinkel, H.; Pomerantz, A.; Prost, S. Genetic biomonitoring and biodiversity assessment using portable sequencing technologies: Current uses and future directions. Genes 2019, 10, 858. [Google Scholar] [CrossRef]
- Sierra, J.; Escobar-Tovar, L.; Leon, P. Plastids: Diving into their diversity, their functions, and their role in plant development. J. Exp. Bot. 2023, 74, 2508–2526. [Google Scholar] [CrossRef]
- Clegg, M.T.; Gaut, B.S.; Learn, G.H., Jr.; Morton, B.R. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 6795–6801. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Jansen, R.K.; Raubeson, L.A.; Boore, J.L.; de Pamphilis, C.W.; Chumley, T.W.; Haberle, R.C.; Wyman, S.K.; Alverson, A.J.; Peery, R.; Herman, S.J.; et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzym. 2005, 395, 348–384. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, A.; Li, X.; Lu, C. The role of chloroplast gene expression in plant responses to environmental stress. Int. J. Mol. Sci. 2020, 21, 6082. [Google Scholar] [CrossRef] [PubMed]
- Yagi, Y.; Shiina, T. Recent advances in the study of chloroplast gene expression and its evolution. Front. Plant Sci. 2014, 5, 61. [Google Scholar] [CrossRef]
- Xiao-Ming, Z.; Junrui, W.; Li, F.; Sha, L.; Hongbo, P.; Lan, Q.; Jing, L.; Yan, S.; Weihua, Q.; Lifang, Z.; et al. Inferring the evolutionary mechanism of the chloroplast genome size by comparing whole-chloroplast genome sequences in seed plants. Sci. Rep. 2017, 7, 1555. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Li, W.-H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef] [PubMed]
- Abdullah; Henriquez, C.L.; Mehmood, F.; Hayat, A.; Sammad, A.; Waseem, S.; Waheed, M.T.; Matthews, P.J.; Croat, T.B.; Poczai, P.; et al. Chloroplast genome evolution in the Dracunculus clade (Aroideae, Araceae). Genomics 2021, 113, 183–192. [Google Scholar] [CrossRef]
- Sun, J.; Wang, Y.; Liu, Y.; Xu, C.; Yuan, Q.; Guo, L.; Huang, L. Evolutionary and phylogenetic aspects of the chloroplast genome of Chaenomeles species. Sci. Rep. 2020, 10, 11466. [Google Scholar] [CrossRef] [PubMed]
- Rohwer, J.G. Toward a phylogenetic classification of the Lauraceae: Evidence from matK sequences. Syst. Bot. 2000, 25, 60. [Google Scholar] [CrossRef]
- Chanderbali, A.S.; Van Der Werff, H.; Renner, S.S. Phylogeny and historical biogeography of Lauraceae: Evidence from the chloroplast and nuclear genomes. Ann. Mo. Bot. Gard. 2001, 88, 104–134. [Google Scholar] [CrossRef]
- Song, Y.; Yu, W.B.; Tan, Y.H.; Jin, J.J.; Wang, B.; Yang, J.B.; Liu, B.; Corlett, R.T. Plastid phylogenomics improve phylogenetic resolution in the Lauraceae. J. Syst. Evol. 2019, 58, 423–439. [Google Scholar] [CrossRef]
- Sampson, F.B.; Berry, P.E. “Laurales”. Encyclopedia britannica 2023. Available online: https://www.britannica.com/plant/Laurales (accessed on 23 March 2024).
- Nishida, S.; van der Werff, H. Are cuticular characters useful in solving generic relationships of problematic species of Lauraceae? Taxon 2007, 56, 1229–1237. [Google Scholar] [CrossRef]
- van der Werff, H.; Nishida, S. Yasunia (Lauraceae), a new genus with two species from Ecuador and Peru. Novon A J. Bot. Nomencl. 2010, 20, 493–502. [Google Scholar] [CrossRef]
- Liu, B.; Yang, Y.; Xie, L.; Zeng, G.; Ma, K. Beilschmiedia turbinata: A newly recognized but dying species of Lauraceae from tropical Asia based on morphological and molecular data. PLoS ONE 2013, 8, e67636. [Google Scholar] [CrossRef]
- IUCN 2024. The IUCN Red List of Threatened Species. Version 2023-1. Available online: https://www.iucnredlist.org/search?query=Beilschmiedia&searchType=species (accessed on 23 March 2024).
- van der Werff, H. Studies in Malagasy Lauraceae II: New taxa. Novon 1996, 6, 463–475. [Google Scholar] [CrossRef]
- Werff, H.v.d. A synopsis of the genus Beilschmiedia (Lauraceae) in Madagascar. Adansonia 2003, 25, 77–92. [Google Scholar]
- Song, Y.; Xia, S.W.; Tan, Y.H.; Yu, W.B.; Yao, X.; Xing, Y.W.; Corlett, R.T. Phylogeny and biogeography of the Cryptocaryeae (Lauraceae). Taxon 2023, 72, 1244–1261. [Google Scholar] [CrossRef]
- Rohwer, J.G.; De Moraes, P.L.R.; Rudolph, B.; Werff, H.V.D. A phylogenetic analysis of the Cryptocarya group (Lauraceae), and relationships of Dahlgrenodendron, Sinopora, Triadodaphne, and Yasunia. Phytotaxa 2014, 158, 111–132. [Google Scholar] [CrossRef]
- Doyle, J.J.; Dickson, E.E. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon 2019, 36, 715–722. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Munch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. The neutral theory of molecular evolution and the world view of the neutralists. Genome 1989, 31, 24–31. [Google Scholar] [CrossRef]
- Yang, Z.; Ma, W.; Yang, X.; Wang, L.; Zhao, T.; Liang, L.; Wang, G.; Ma, Q. Plastome phylogenomics provide new perspective into the phylogeny and evolution of Betulaceae (Fagales). BMC Plant Biol. 2022, 22, 611. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, L.; Teixeira da Silva, J.A.; Yu, X. The plastome reveals new insights into the evolutionary and domestication history of peonies in East Asia. BMC Plant Biol. 2023, 23, 243. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Du, Y.; Xie, L.; Jin, X.; Zhang, Z.; Yang, M. Comparative plastome genomics and phylogenetic relationships of the genus Trollius. Front. Plant Sci. 2023, 14, 1293091. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, B.; Davis, C.C.; Yang, Y. Plastome phylogenomics, systematics, and divergence time estimation of the Beilschmiedia group (Lauraceae). Mol. Phylogenet Evol. 2020, 151, 106901. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Tan, Y.; Zhou, X.; Song, Y.; Xin, P. The complete plastid genome sequences of the Belian (Eusideroxylon zwageri): Comparative aanalysis and phylogenetic relationships with other Magnoliids. Forests 2023, 14, 2443. [Google Scholar] [CrossRef]
- Song, Y.; Dong, W.; Liu, B.; Xu, C.; Yao, X.; Gao, J.; Corlett, R.T. Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Front. Plant Sci. 2015, 6, 662. [Google Scholar] [CrossRef] [PubMed]
- Marechal, A.; Brisson, N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010, 186, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.H.; Kong, B.L.; Siu, T.Y.; Wu, H.Y.; But, G.W.; Shaw, P.C.; Lau, D.T. Complete chloroplast genomes of Asparagus aethiopicus L., A. densiflorus (Kunth) Jessop M‘yers’, and A. cochinchinensis (Lour.) Merr.: Comparative and phylogenetic analysis with congenerics. PLoS ONE 2022, 17, e0266376. [Google Scholar] [CrossRef]
- Guo, L.; Guo, S.; Xu, J.; He, L.; Carlson, J.E.; Hou, X. Phylogenetic analysis based on chloroplast genome uncover evolutionary relationship of all the nine species and six cultivars of tree peony. Ind. Crops Prod. 2020, 153, 112567. [Google Scholar] [CrossRef]
- Loewe, L.; Hill, W.G. The population genetics of mutations: Good, bad and indifferent. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 1153–1167. [Google Scholar] [CrossRef] [PubMed]
- Castellano, D.; Eyre-Walker, A.; Munch, K. Impact of mutation rate and selection at linked sites on DNA variation across the genomes of humans and other homininae. Genome Biol. Evol. 2020, 12, 3550–3561. [Google Scholar] [CrossRef]
- Clancy, S. Genetic mutation. Nat. Educ. 2008, 1, 187. [Google Scholar]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Worberg, A.; Quandt, D.; Barniske, A.-M.; Löhne, C.; Hilu, K.W.; Borsch, T. Phylogeny of basal eudicots: Insights from non-coding and rapidly evolving DNA. Org. Divers. Evol. 2007, 7, 55–77. [Google Scholar] [CrossRef]
- Ahmed, I.; Matthews, P.J.; Biggs, P.J.; Naeem, M.; McLenachan, P.A.; Lockhart, P.J. Identification of chloroplast genome loci suitable for high-resolution phylogeographic studies of Colocasia esculenta (L.) Schott (Araceae) and closely related taxa. Mol. Ecol. Resour. 2013, 13, 929–937. [Google Scholar] [CrossRef]
- Xu, S.; Stapley, J.; Gablenz, S.; Boyer, J.; Appenroth, K.J.; Sree, K.S.; Gershenzon, J.; Widmer, A.; Huber, M. Low genetic variation is associated with low mutation rate in the giant duckweed. Nat. Commun. 2019, 10, 1243. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, M.J.; Dugan, J.M.; Shafer, R.W. Synonymous-non-synonymous mutation rates between sequences containing ambiguous nucleotides (Syn-SCAN). Bioinformatics 2002, 18, 886–887. [Google Scholar] [CrossRef] [PubMed]
- Pond, S.K.; Muse, S.V. Site-to-site variation of synonymous substitution rates. Mol. Biol. Evol. 2005, 22, 2375–2385. [Google Scholar] [CrossRef] [PubMed]
- Lee-Yaw, J.A.; Grassa, C.J.; Joly, S.; Andrew, R.L.; Rieseberg, L.H. An evaluation of alternative explanations for widespread cytonuclear discordance in annual sunflowers (Helianthus). New Phytol. 2019, 221, 515–526. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; dePamphilis, C.W.; Muller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Drescher, A.; Ruf, S.; Calsa, T., Jr.; Carrer, H.; Bock, R. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 2000, 22, 97–104. [Google Scholar] [CrossRef] [PubMed]






| Beilschmiedia moratii | Potameia microphylla | Beilschmiedia pierreana | |
|---|---|---|---|
| Beilschmiedia moratii | - | 0.00283 | 0.00459 |
| Potameia microphylla | 0.00283 | - | 0.00405 |
| Beilschmiedia pierreana | 0.00459 | 0.00405 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, W.; Zhang, H.; Li, Q.; Cao, Z.; Song, Y.; Xin, P. Complete Plastid Genome Sequences of Three Tropical African Beilschmiediineae Trees (Lauraceae: Crytocaryeae). Forests 2024, 15, 832. https://doi.org/10.3390/f15050832
Zhu W, Zhang H, Li Q, Cao Z, Song Y, Xin P. Complete Plastid Genome Sequences of Three Tropical African Beilschmiediineae Trees (Lauraceae: Crytocaryeae). Forests. 2024; 15(5):832. https://doi.org/10.3390/f15050832
Chicago/Turabian StyleZhu, Wen, Haorong Zhang, Qishao Li, Zhengying Cao, Yu Song, and Peiyao Xin. 2024. "Complete Plastid Genome Sequences of Three Tropical African Beilschmiediineae Trees (Lauraceae: Crytocaryeae)" Forests 15, no. 5: 832. https://doi.org/10.3390/f15050832




