Enhancing Cellular Uptake of Native Proteins through Bio-Orthogonal Conjugation with Chemically Synthesized Cell-Penetrating Peptides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture Maintenance
2.2. Reporter Plasmid Design and Cloning
2.3. Peptide Synthesis
2.4. Cu-Catalyzed Azide–Alkyne Cycloaddition for Intein-Activated CPP NF55 Synthesis
2.5. Transfection and Fusion Protein Production
2.6. Flow Cytometry
2.7. Confocal Microscopy
3. Results
3.1. Design of Nanoentity for Intracellular Protein Delivery
3.2. Split EGFP Assay for Bioconjugation Method Validation in Mammalian Cells
3.3. Synthesis of the Nanoentity Substrate Compounds
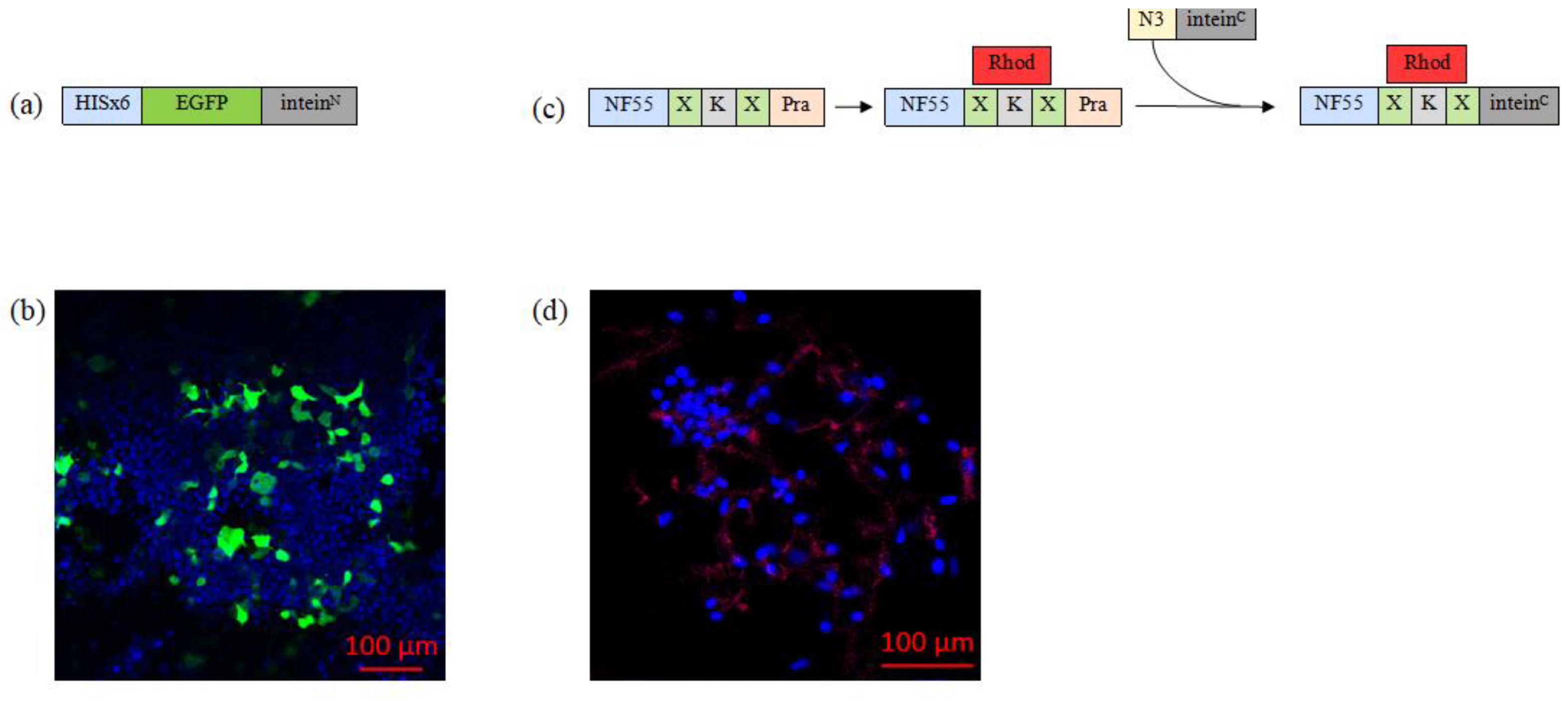
3.3.1. Substrate 1: Biosynthesis of EGFP–InteinN Fusion Protein
3.3.2. Substrate 2: Chemical Synthesis of Inteinc-activated NF55 Peptide
- (1)
- NF55, with extended C-terminus to include propargylglycine for later click reaction, was synthesized and labeled with carboxytetramethylrhodamine (Rhod).
- (2)
- InteinC peptide was designed to contain azide group.
- (3)
- The fluorolabeled NF55 and azide–inteinC peptide were then conjugated through copper-catalyzed azide–alkyne click reaction. The successful formation of the nanoentity substrate was confirmed via UPLC (Figure S3).
3.4. Assembly of the full Nanoentity and Protein Transduction in Mammalian Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vargason, A.M.; Anselmo, A.C.; Mitragotri, S. The evolution of commercial drug delivery technologies. Nat. Biomed. Eng. 2021, 5, 951–967. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Choi, D.W.; Kim, H.N.; Park, C.G.; Lee, W.; Park, H.H. Protein-based nanoparticles as drug delivery systems. Pharmaceutics 2020, 12, 604. [Google Scholar] [CrossRef] [PubMed]
- Moncalvo, F.; Martinez Espinoza, M.I.; Cellesi, F. Nanosized Delivery Systems for Therapeutic Proteins: Clinically Validated Technologies and Advanced Development Strategies. Front. Bioeng. Biotechnol. 2020, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Kintzing, J.R.; Filsinger Interrante, M.V.; Cochran, J.R. Emerging strategies for developing next-generation protein therapeutics for cancer treatment. Trends Pharmacol. Sci. 2016, 37, 993–1008. [Google Scholar] [CrossRef] [PubMed]
- Lagassé, H.A.D.; Alexaki, A.; Simhadri, V.L.; Katagiri, N.H.; Jankowski, W.; Sauna, Z.E.; Kimchi-Sarfaty, C. Recent advances in (therapeutic protein) drug development. F1000Research 2017, 6, 113. [Google Scholar] [CrossRef] [PubMed]
- Amer, M.H. Gene therapy for cancer: Present status and future perspective. Mol. Cell. Ther. 2014, 2, 27. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Parveen, R.; Chatterji, B.P. Toxicology of nanoparticles in drug delivery. Curr. Pathobiol. Rep. 2021, 9, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, T.; de Zafra, C.; Kim, A.; Gelzleichter, T.R. Overview of biopharmaceuticals and comparison with small-molecule drug development. In Nonclinical Development of Novel Biologics, Biosimilars, Vaccines and Specialty Biologics; Elsevier: Amsterdam, The Netherlands, 2013; pp. 3–33. [Google Scholar]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Steichen, J.M.; Kuchinskas, M.; Keshwani, M.M.; Yang, J.; Adams, J.A.; Taylor, S.S. Structural basis for the regulation of protein kinase A by activation loop phosphorylation. J. Biol. Chem. 2012, 287, 14672–14680. [Google Scholar] [CrossRef]
- Patil, N.A.; Tailhades, J.; Hughes, R.A.; Separovic, F.; Wade, J.D.; Hossain, M.A. Cellular disulfide bond formation in bioactive peptides and proteins. Int. J. Mol. Sci. 2015, 16, 1791–1805. [Google Scholar] [CrossRef]
- Levine, Z.G.; Potter, S.C.; Joiner, C.M.; Fei, G.Q.; Nabet, B.; Sonnett, M.; Zachara, N.E.; Gray, N.S.; Paulo, J.A.; Walker, S. Mammalian cell proliferation requires noncatalytic functions of O-GlcNAc transferase. Proc. Natl. Acad. Sci. USA 2021, 118, e2016778118. [Google Scholar] [CrossRef]
- Ron, D. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 2002, 110, 1383–1388. [Google Scholar] [CrossRef] [PubMed]
- Milletti, F. Cell-penetrating peptides: Classes, origin, and current landscape. Drug Discov. Today 2012, 17, 850–860. [Google Scholar] [CrossRef]
- Langel, Ü. Therapeutic Potential of CPPs. In CPP, Cell-Penetrating Peptides; Springer: Singapore, 2019; pp. 409–461. [Google Scholar]
- Chatterjee, S.; Kon, E.; Sharma, P.; Peer, D. Endosomal escape: A bottleneck for LNP-mediated therapeutics. Proc. Natl. Acad. Sci. USA 2024, 121, e2307800120. [Google Scholar] [CrossRef] [PubMed]
- Dowdy, S.F. Endosomal escape of RNA therapeutics: How do we solve this rate-limiting problem? RNA 2023, 29, 396–401. [Google Scholar] [CrossRef]
- Kristensen, M.; Birch, D.; Mørck Nielsen, H. Applications and challenges for use of cell-penetrating peptides as delivery vectors for peptide and protein cargos. Int. J. Mol. Sci. 2016, 17, 185. [Google Scholar] [CrossRef]
- Guterstam, P.; Madani, F.; Hirose, H.; Takeuchi, T.; Futaki, S.; El Andaloussi, S.; Gräslund, A.; Langel, Ü. Elucidating cell-penetrating peptide mechanisms of action for membrane interaction, cellular uptake, and translocation utilizing the hydrophobic counter-anion pyrenebutyrate. Biochim. Biophys. Acta (BBA)-Biomembr. 2009, 1788, 2509–2517. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Tan, S.; Bao, Y.; Zhang, Z. Enhanced tumor therapy via drug co-delivery and in situ vascular-promoting strategy. J. Control. Release 2017, 258, 108–120. [Google Scholar] [CrossRef]
- Miwa, T.; Tachii, K.; Wei, F.-Y.; Kaitsuka, T.; Tomizawa, K. Intranasal drug delivery into mouse nasal mucosa and brain utilizing arginine-rich cell-penetrating peptide-mediated protein transduction. Int. J. Pept. Res. Ther. 2020, 26, 1643–1650. [Google Scholar] [CrossRef]
- Ray, M.; Lee, Y.-W.; Scaletti, F.; Yu, R.; Rotello, V.M. Intracellular delivery of proteins by nanocarriers. Nanomedicine 2017, 12, 941–952. [Google Scholar] [CrossRef]
- Munyendo, W.L.; Lv, H.; Benza-Ingoula, H.; Baraza, L.D.; Zhou, J. Cell penetrating peptides in the delivery of biopharmaceuticals. Biomolecules 2012, 2, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Bolhassani, A.; Jafarzade, B.S.; Mardani, G. In vitro and in vivo delivery of therapeutic proteins using cell penetrating peptides. Peptides 2017, 87, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.F.L.; Wallabregue, A.L.D.; Franz, L.; Hackenberger, C.P.R. Targeted subcellular protein delivery using cleavable cyclic cell-penetrating peptides. Bioconjugate Chem. 2019, 30, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Kardani, K.; Milani, A.; HShabani, S.; Bolhassani, A. Cell penetrating peptides: The potent multi-cargo intracellular carriers. Expert Opin. Drug Deliv. 2019, 16, 1227–1258. [Google Scholar] [CrossRef] [PubMed]
- Langel, Ü. Cell-penetrating peptides and transportan. Pharmaceutics 2021, 13, 987. [Google Scholar] [CrossRef]
- Freimann, K.; Arukuusk, P.; Kurrikoff, K.; Vasconcelos, L.D.F.; Veiman, K.-L.; Uusna, J.; Margus, H.; Garcia-Sosa, A.T.; Pooga, M.; Langel, Ü. Optimization of in vivo DNA delivery with NickFect peptide vectors. J. Control. Release 2016, 241, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-X.; Wei, Y.; Zhong, R.; Li, L.; Pang, H.-B. Transportan peptide stimulates the nanomaterial internalization into mammalian cells in the bystander manner through macropinocytosis. Pharmaceutics 2021, 13, 552. [Google Scholar] [CrossRef] [PubMed]
- Pärnaste, L.; Arukuusk, P.; Zagato, E.; Braeckmans, K.; Langel, Ü. Methods to follow intracellular trafficking of cell-penetrating peptides. J. Drug Target. 2016, 24, 508–519. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, S.; Terhorst, T.M.E.; Singh, R.K.; Kümmel, D.; Pietrokovski, S.; Mootz, H.D. Biochemical and structural characterization of an unusual and naturally split class 3 intein. Chembiochem 2021, 22, 364–373. [Google Scholar] [CrossRef]
- Antos, J.M.; Truttmann, M.C.; Ploegh, H.L. Recent advances in sortase-catalyzed ligation methodology. Curr. Opin. Struct. Biol. 2016, 38, 111–118. [Google Scholar] [CrossRef]
- McKay, C.S.; Finn, M.G. Click chemistry in complex mixtures: Bioorthogonal bioconjugation. Chem. Biol. 2014, 21, 1075–1101. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P.E.; Kent, S.B. Synthesis of native proteins by chemical ligation. Annu. Rev. Biochem. 2000, 69, 923–960. [Google Scholar] [CrossRef] [PubMed]
- Reguera, L.; Méndez, Y.; Humpierre, A.R.; Valdés, O.; Rivera, D.G. Multicomponent reactions in ligation and bioconjugation chemistry. Acc. Chem. Res. 2018, 51, 1475–1486. [Google Scholar] [CrossRef] [PubMed]
- Agouridas, V.; El Mahdi, O.; Diemer, V.; Cargoët, M.; Monbaliu, J.-C.M.; Melnyk, O. Native chemical ligation and extended methods: Mechanisms, catalysis, scope, and limitations. Chem. Rev. 2019, 119, 7328–7443. [Google Scholar] [CrossRef] [PubMed]
- Aranko, A.S.; Züger, S.; Buchinger, E.; Iwaï, H. In vivo and in vitro protein ligation by naturally occurring and engineered split DnaE inteins. PLoS ONE 2009, 4, e5185. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Siu, K.-H.; Raeeszadeh-Sarmazdeh, M.; Sun, Q.; Chen, Q.; Chen, W. Bioengineering strategies to generate artificial protein complexes. Biotechnol. Bioeng. 2015, 112, 1495–1505. [Google Scholar] [CrossRef] [PubMed]
- Dhar, T.; Kurpiers, T.; Mootz, H.D. Extending the scope of site-specific cysteine bioconjugation by appending a prelabeled cysteine tag to proteins using protein trans-splicing. Methods Mol. Biol. 2011, 751, 131–142. [Google Scholar] [PubMed]
- Romero-Casañas, A.; Gordo, V.; Castro, J.; Ribó, M. Protein splicing: From the foundations to the development of biotechnological applications. Methods Mol. Biol. 2020, 2133, 15–29. [Google Scholar] [PubMed]
- David, Y.; Vila-Perelló, M.; Verma, S.; Muir, T.W. Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins. Nat. Chem. 2015, 7, 394–402. [Google Scholar] [CrossRef]
- Presolski, S.I.; Hong, V.P.; Finn, M.G. Copper-catalyzed azide-alkyne click chemistry for bioconjugation. Curr. Protoc. Chem. Biol. 2011, 3, 153–162. [Google Scholar] [CrossRef]
- Porosk, L.; Nebogatova, J.; Härk, H.H.; Vunk, B.; Arukuusk, P.; Toots, U.; Ustav, M.; Langel, Ü.; Kurrikoff, K. Predicting transiently expressed protein yields: Comparison of transfection methods in CHO and HEK293. Pharmaceutics 2022, 14, 1949. [Google Scholar] [CrossRef] [PubMed]
- Härk, H.H.; Porosk, L.; de Mello, L.R.; Arukuusk, P.; da Silva, E.R.; Kurrikoff, K. Modification of the linker amino acid in the cell-penetrating peptide NickFect55 leads to enhanced pDNA transfection for in vivo applications. Pharmaceutics 2023, 15, 883. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef] [PubMed]
- Marvin Documentation—Marvin 2.8.0 Documentation. Available online: http://sdss-marvin.readthedocs.io/en/stable/ (accessed on 3 April 2024).
- Shah, N.H.; Dann, G.P.; Vila-Perelló, M.; Liu, Z.; Muir, T.W. Ultrafast protein splicing is common among cyanobacterial split inteins: Implications for protein engineering. J. Am. Chem. Soc. 2012, 134, 11338–11341. [Google Scholar] [CrossRef] [PubMed]



| Name | Advantages | Limitations |
|---|---|---|
| Click chemistry [32,33] | Highly efficient, specific, and bio-orthogonal | Requires specific reactive groups and is not universally suitable for all biomolecules |
| Sortase-mediated ligation [32] | Site-specific, efficient, and allows for the incorporation of synthetic peptides into proteins | Limited to those containing a specific motif, potentially requires optimization for various substrates, and has limited activity and stability |
| Native chemical ligation [34,35,36] | Compatible with a broad range of biomolecules and suitable for large proteins | Necessitates a cysteine residue at the ligation site, and reaction rates may be slow for certain substrates |
| Protein-trans splicing (NCL analog) [37,38,39,40] | Rapid and can occur in a biological environment without affecting the higher-order structure of the protein | Limited to those containing a specific intein sequence |
| Name | Description | Size |
|---|---|---|
| EGFP | Reporter protein component of the nanoentity. Naturally occurring fluorescent protein derived from the jellyfish Aequorea ictoria. | 714 amino acid residues |
| NickFect 55 (NF55) | Cell-penetrating peptide/vehicle of the nanoentity. Analog of CPP Transportan 10. Stearoyl-AGYLLGO*INLKALAALAKAIL-NH2; O*—synthesis continued from the side-chain instead of the alpha-amino group. | 21 amino acid residues |
| InteinN | N-terminal split intein fragment. Origin from DNA polymerase III subunit alpha; Anabaena variabilis ATCC 29413. | 80 amino acid residues |
| InteinC | C-terminal split intein fragment. Origin from DNA polymerase III subunit alpha; Nostoc sp. ATCC 53789. | 39 amino acid residues |
| Name | EGFP Fragment | Spilt Intein Fragment | Control Reporter | |
|---|---|---|---|---|
| First set | p_egfpN_intN | EGFP N-terminal fragment | inteinN | mCherry (red) |
| p_intC_egfpC | EGFP N-terminal fragment | inteinC | SBFP (blue) | |
| Second set | p_egfp1-10_intN | EGFP 1–10 domains | inteinN | mCherry (red) |
| p_intC_egfp11 | EGFP 11 domain | inteinC | SBFP (blue) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nebogatova, J.; Porosk, L.; Härk, H.H.; Kurrikoff, K. Enhancing Cellular Uptake of Native Proteins through Bio-Orthogonal Conjugation with Chemically Synthesized Cell-Penetrating Peptides. Pharmaceutics 2024, 16, 617. https://doi.org/10.3390/pharmaceutics16050617
Nebogatova J, Porosk L, Härk HH, Kurrikoff K. Enhancing Cellular Uptake of Native Proteins through Bio-Orthogonal Conjugation with Chemically Synthesized Cell-Penetrating Peptides. Pharmaceutics. 2024; 16(5):617. https://doi.org/10.3390/pharmaceutics16050617
Chicago/Turabian StyleNebogatova, Jekaterina, Ly Porosk, Heleri Heike Härk, and Kaido Kurrikoff. 2024. "Enhancing Cellular Uptake of Native Proteins through Bio-Orthogonal Conjugation with Chemically Synthesized Cell-Penetrating Peptides" Pharmaceutics 16, no. 5: 617. https://doi.org/10.3390/pharmaceutics16050617






