Chitin Deacetylase Homologous Gene cda Contributes to Development and Aflatoxin Synthesis in Aspergillus flavus
Abstract
:1. Introduction
2. Results
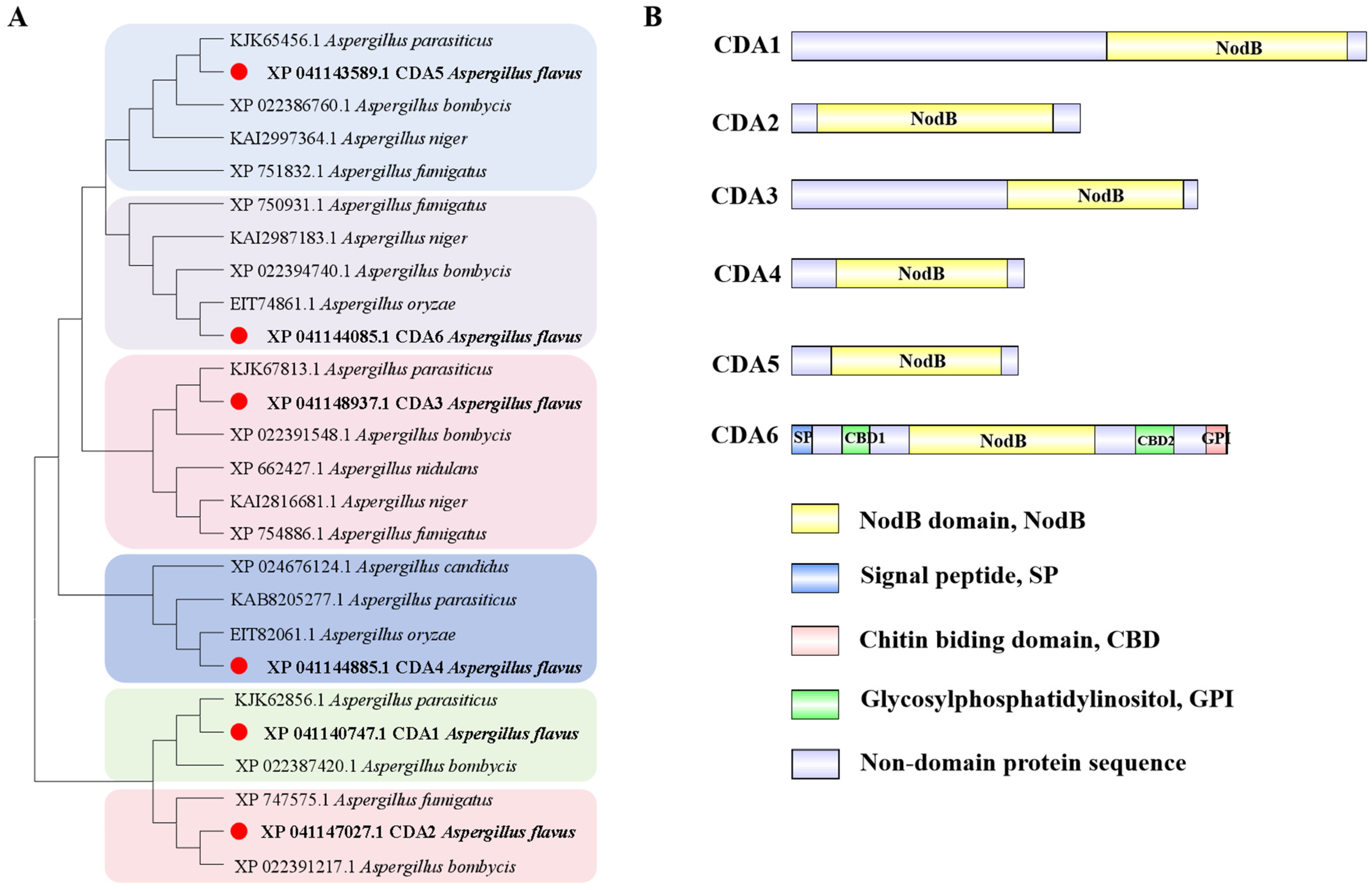
2.1. Bioinformatics Analysis of CDA Family in A. flavus
2.2. Quantitative Analysis of Chitin in A. flavus
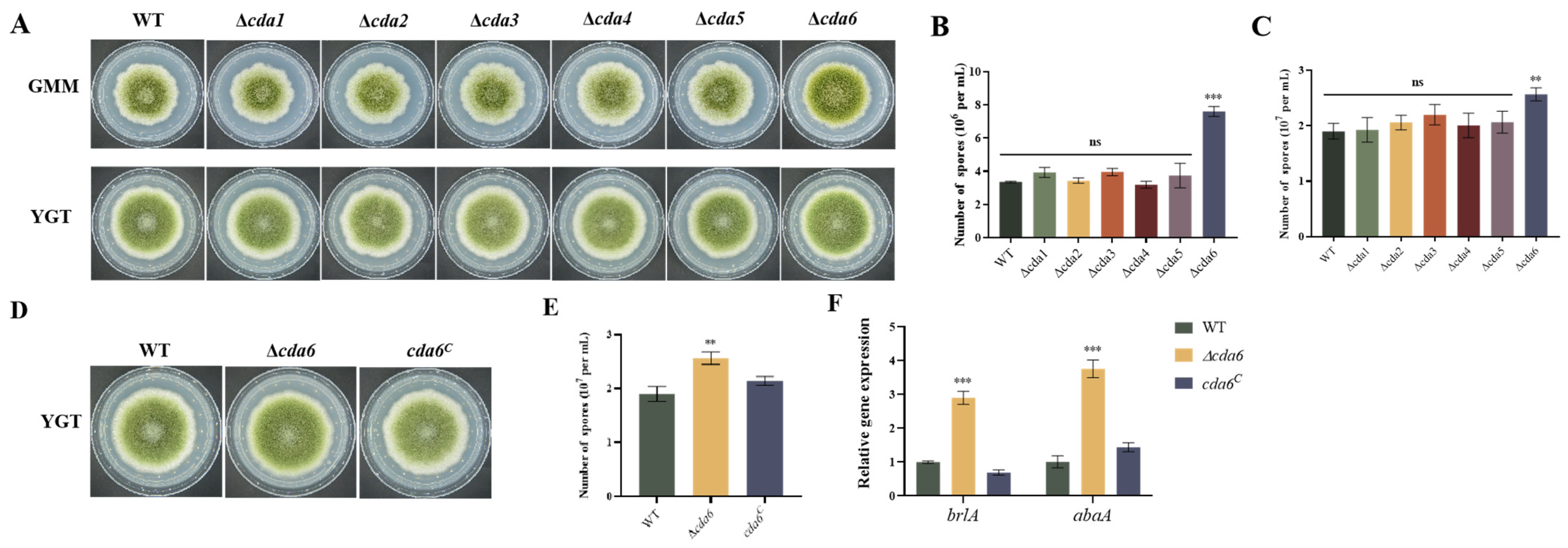
2.3. Involvement of CDA Members in Regulating Conidial Formation in A. flavus
2.4. Fluorescence Localization of CDA6 Protein in A. flavus
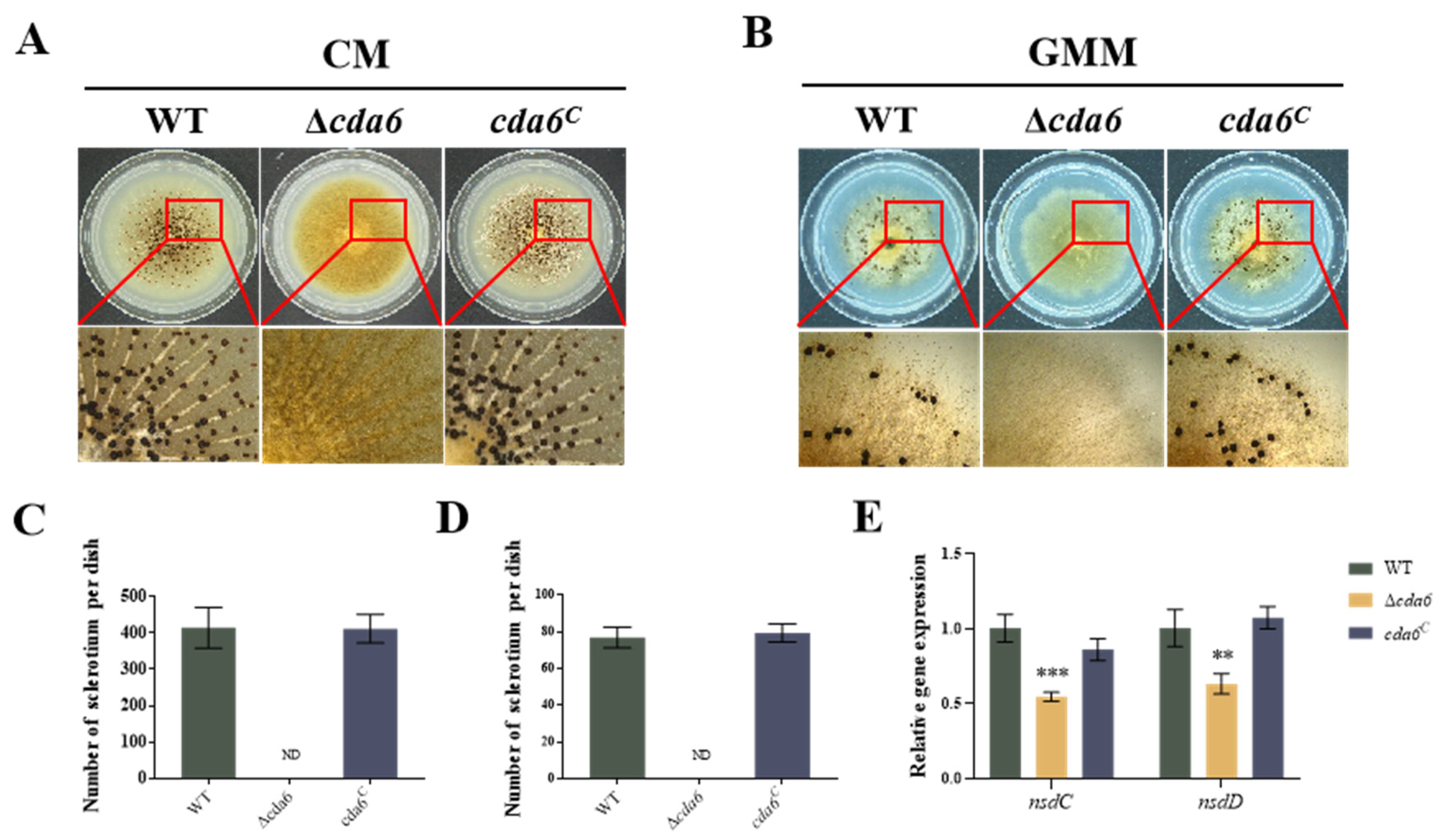
2.5. cda6 Affects the Sclerotia Formation in A. flavus
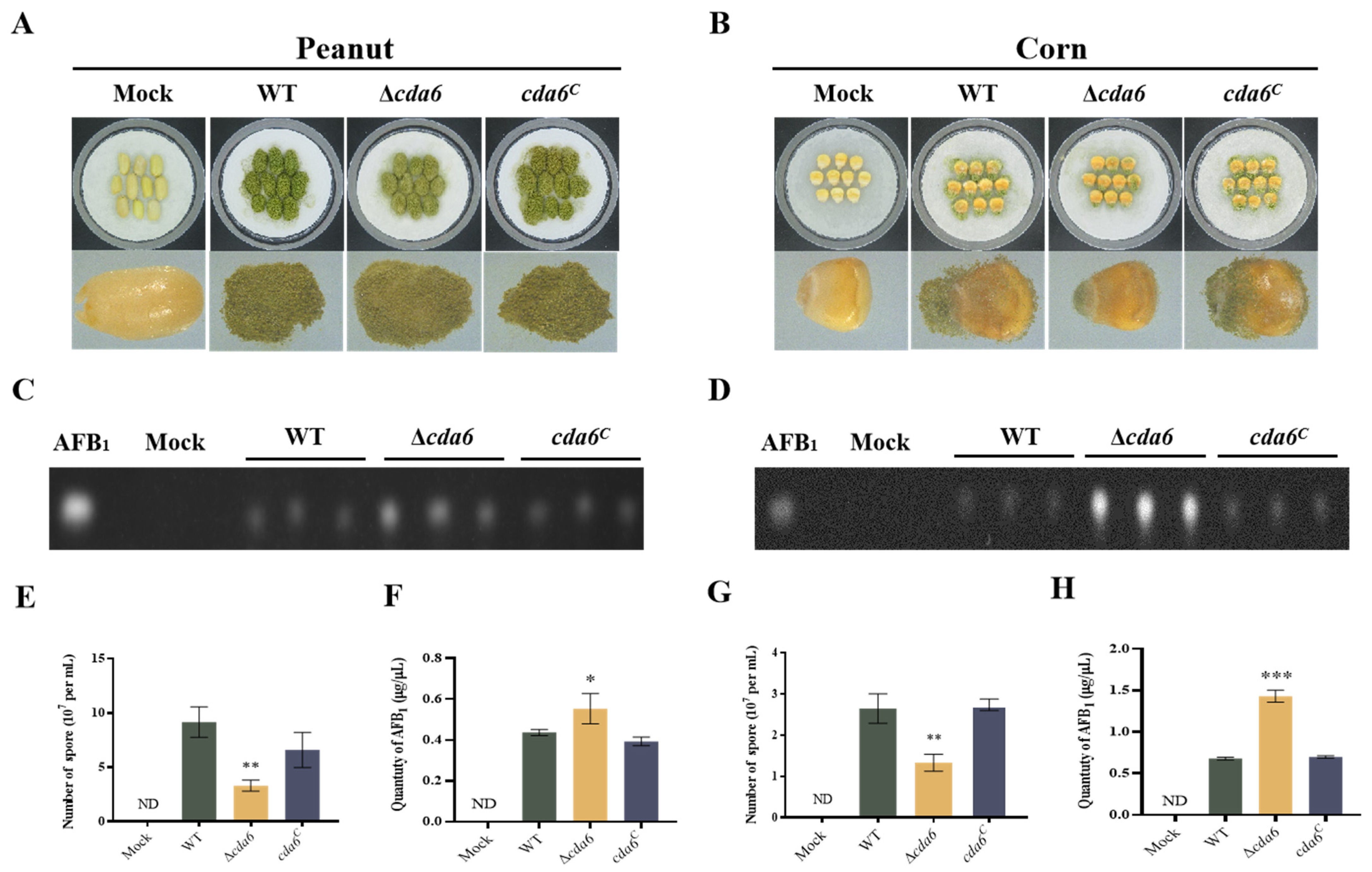
2.6. Effect of cda6 Gene on Aflatoxin Biosynthesis
2.7. cda6 Gene Deletion Affects A. flavus Pathogenicity
2.8. The Role of cda6 Gene in the Stress Response
2.9. Functional Studies of Structural Domains and Signal Peptide of CDA6 in A. flavus
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Strains and Culture Conditions
5.2. Sequence Analysis
5.3. Construction of Mutant Strains
5.4. Calcofluor White (CFW) Staining
5.5. Chitin Quantification
5.6. Extraction and Determination of Aflatoxin
5.7. qPCR Analysis
5.8. Seed Infections Assay
5.9. Stress Response Analysis
5.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klich, M.A. Aspergillus flavus: The major producer of aflatoxin. Mol. Plant Pathol. 2007, 8, 713–722. [Google Scholar] [CrossRef]
- Probst, C.; Schulthess, F.; Cotty, P.J. Impact of Aspergillus section Flavi community structure on the development of lethal levels of aflatoxins in Kenyan maize (Zea mays). J. Appl. Microbiol. 2010, 108, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Lewis, L.; Onsongo, M.; Njapau, H.; Schurz-Rogers, H.; Luber, G.; Kieszak, S.; Nyamongo, J.; Backer, L.; Dahiye, A.M.; Misore, A.; et al. Aflatoxin contamination of commercial maize products during an outbreak of acute aflatoxicosis in eastern and central Kenya. Environ. Health Perspect. 2005, 113, 1763–1767. [Google Scholar] [CrossRef] [PubMed]
- Hedayati, M.T.; Pasqualotto, A.C.; Warn, P.A.; Bowyer, P.; Denning, D.W. Aspergillus flavus: Human pathogen, allergen and mycotoxin producer. Microbiology 2007, 153, 1677–1692. [Google Scholar] [CrossRef] [PubMed]
- Rudramurthy, S.M.; Paul, R.A.; Chakrabarti, A.; Mouton, J.W.; Meis, J.F. Invasive Aspergillosis by Aspergillus flavus: Epidemiology, diagnosis, antifungal resistance, and management. J. Fungi. 2019, 5, 55. [Google Scholar] [CrossRef]
- Caceres, I.; Khoury, A.A.; Khoury, R.E.; Lorber, S.; Oswald, I.P.; Khoury, A.E.; Atoui, A.; Puel, O.; Bailly, J.D. Aflatoxin biosynthesis and genetic regulation: A review. Toxins 2020, 12, 150. [Google Scholar] [CrossRef]
- Latgé, J.P. The cell wall: A carbohydrate armour for the fungal cell. Mol. Microbiol. 2007, 66, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. Bioessays 2006, 28, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Latge, J.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 1128. [Google Scholar] [CrossRef]
- Perlin, D.S. Cell wall-modifying antifungal drugs. Curr. Top. Microbiol. 2020, 425, 255–275. [Google Scholar] [CrossRef]
- Munro, C.A.; Winter, K.; Buchan, A.; Henry, K.; Becker, J.M.; Brown, A.J.P.; Bulawa, C.E.; Gow, N.A.R. Chs1 of candida albicans is an essential chitin synthase required for synthesis of the septum and for cell integrity. Mol. Microbiol. 2001, 39, 1414–1426. [Google Scholar] [CrossRef]
- Tsigos, I.; Martinou, A.; Kafetzopoulos, D.; Bouriotis, V. Chitin deacetylases: New, versatile tools in biotechnology. Trends Biotechnol. 2000, 18, 305–312. [Google Scholar] [CrossRef]
- Christodoulidou, A.; Briza, P.; Ellinger, A.; Bouriotis, V. Yeast ascospore wall assembly requires two chitin deacetylase isozymes. FEBS Lett. 1999, 460, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Christodoulidou, A.; Bouriotis, V.; Thireos, G. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 31420–31425. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.G.; Specht, C.A.; Donlin, M.J.; Lodge, J.K. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell. 2007, 6, 855–867. [Google Scholar] [CrossRef]
- Upadhya, R.; Baker, L.G.; Lam, W.C.; Specht, C.A.; Donlin, M.J.; Lodge, J.K. Cryptococcus neoformans Cda1 and its chitin deacetylase activity are required for fungal pathogenesis. mBio 2018, 9, e02087-18. [Google Scholar] [CrossRef]
- Dai, M.D.; Wu, M.; Li, Y.; Su, Z.Z.; Lin, F.C.; Liu, X.H. The chitin deacetylase PoCda7 is involved in the pathogenicity of Pyricularia oryzae. Microbiol. Res. 2021, 248, 126749. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Zhao, X.; Lü, Y.; Jin, C. Chitin deacetylases Cod4 and Cod7 are involved in polar growth of Aspergillus fumigatus. Microbiologyopen 2020, 9, e00943. [Google Scholar] [CrossRef]
- Mouyna, I.; Dellière, S.; Beauvais, A.; Gravelat, F.; Snarr, B.; Lehoux, M.; Zacharias, C.; Sun, Y.; de Jesus Carrion, S.; Pearlman, E.; et al. What are the functions of chitin deacetylases in Aspergillus fumigatus? Front. Cell. Infect. Microbiol. 2020, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- John, M.; Röhrig, H.; Schmidt, J.; Wieneke, U.; Schell, J. Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc. Natl. Acad. Sci. USA 1993, 90, 625–629. [Google Scholar] [CrossRef]
- Lee, S.; Kang, H.A.; Eyun, S.I. Evolutionary analysis and protein family classification of chitin deacetylases in Cryptococcus neoformans. J. Microbiol. 2020, 58, 805–811. [Google Scholar] [CrossRef]
- Müller, G.A.; Tschöp, M.H.; Müller, T.D. Chip-based sensing of the intercellular transfer of cell surface proteins: Regulation by the metabolic state. Biomedicine 2021, 9, 1452. [Google Scholar] [CrossRef]
- Lima, S.L.; Colombo, A.L.; Junior, J.N.D.A. Fungal cell wall: Emerging antifungals and drug resistance. Front. Microbiol. 2019, 10, 2573. [Google Scholar] [CrossRef]
- Kamakura, T.; Yamaguchi, S.; Saitoh, K.; Teraoka, T.; Yamaguchi, I. A novel gene, CBP1, encoding a putative extracellular chitin-binding protein, may play an important role in the hydrophobic surface sensing of Magnaporthe grisea during appressorium differentiation. Mol. Plant Microbe Interact. 2002, 15, 437–444. [Google Scholar] [CrossRef]
- Rizzi, Y.S.; Happel, P.; Lenz, S.; Urs, M.J.; Bonin, M.; Cord-Landwehr, S.; Singh, R.; Moerschbacher, B.M.; Kahmann, R. Chitosan and chitin deacetylase activity are necessary for development and virulence of Ustilago maydis. mBio 2021, 12, e03419-20. [Google Scholar] [CrossRef] [PubMed]
- Delgado, J.; Owens, R.A.; Doyle, S.; Asensio, M.A.; Núñez, F. Increased chitin biosynthesis contributes to the resistance of Penicillium polonicum against the antifungal protein PgAFP. Appl. Microbiol. Biotechnol. 2016, 100, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Yago, J.I.; Lin, C.H.; Chung, K.R. The SLT2 mitogen-activated protein kinase-mediated signalling pathway governs conidiation, morphogenesis, fungal virulence and production of toxin and melanin in the tangerine pathotype of Alternaria alternata. Mol. Plant Pathol. 2011, 12, 653–665. [Google Scholar] [CrossRef]
- Yu, J.H. Regulation of development in Aspergillus nidulans and Aspergillus fumigatus. Mycobiology 2010, 38, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; Tanaka, K.; Matsuda, H.; Kawamukai, M. cda1+, encoding chitin deacetylase is required for proper spore formation in Schizosaccharomyces pombe. FEBS Lett. 2005, 579, 2737–2743. [Google Scholar] [CrossRef]
- Sun, X.; Liu, D.; Wang, Y.; Ma, A. Biogenesis of macrofungal sclerotia: Influencing factors and molecular mechanisms. Appl. Microbiol. Biotechnol. 2020, 104, 4227–4234. [Google Scholar] [CrossRef]
- Dyer, P.S.; O’Gorman, C.M. Sexual development and cryptic sexuality in fungi: Insights from Aspergillus species. FEMS Microbiol. Rev. 2012, 36, 165–192. [Google Scholar] [CrossRef]
- Gao, X.D.; Katsumoto, T.; Onodera, K. Purification and characterization of chitin deacetylase from Absidia coerulea. J. Biochem. 1995, 117, 257–263. [Google Scholar] [CrossRef]
- Kafetzopoulos, D.; Martinou, A.; Bouriotis, V. Bioconversion of chitin to chitosan: Purification and characterization of chitin deacetylase from Mucor rouxii. Proc. Natl. Acad. Sci. USA 1993, 90, 2564–2568. [Google Scholar] [CrossRef] [PubMed]
- Alfonso, C.; Nuero, O.M.; Santamaría, F.; Reyes, F. Purification of a heat-stable chitin deacetylase from Aspergillus nidulans and its role in cell wall degradation. Curr. Microbiol. 1995, 30, 49–54. [Google Scholar] [CrossRef]
- Tokuyasu, K.; Mitsutomi, M.; Yamaguchi, I.; Hayashi, K.; Mori, Y. Recognition of chitooligosaccharides and their N-acetyl groups by putative subsites of chitin deacetylase from a deuteromycete, Colletotrichum lindemuthianum. Biochemistry 2000, 39, 8837–8843. [Google Scholar] [CrossRef]
- Rushing, B.R.; Selim, M.I. Aflatoxin B1: A review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem. Toxicol. 2019, 124, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Amaike, S.; Keller, N.P. Aspergillus flavus. Annu. Rev. Phytopathol. 2011, 49, 107–133. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.; Wilkinson, J.; Zablotowicz, R.; Accinelli, C.; Abel, C.; Bruns, H.; Weaver, M. Ecology of Aspergillus flavus, regulation of aflatoxin production, and management strategies to reduce aflatoxin contamination of corn. Toxin Rev. 2009, 28, 142–153. [Google Scholar] [CrossRef]
- Prasad, R.; Shah, A.H.; Rawal, M.K. Antifungals: Mechanism of action and drug resistance. Adv. Exp. Med. Biol. 2016, 892, 327–349. [Google Scholar] [CrossRef]
- Costa-de-Oliveira, S.; Silva, A.P.; Miranda, I.M.; Salvador, A.; Azevedo, M.M.; Munro, C.A.; Rodrigues, A.G.; Pina-Vaz, C. Determination of chitin content in fungal cell wall: An alternative flow cytometric method. Cytom. A 2013, 83, 324–328. [Google Scholar] [CrossRef]
- Plaine, A.; Walker, L.; Da Costa, G.; Mora-Montes, H.M.; McKinnon, A.; Gow, N.A.; Gaillardin, C.; Munro, C.A.; Richard, M.L. Functional analysis of Candida albicans GPI-anchored proteins: Roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 2008, 45, 1404–1414. [Google Scholar] [CrossRef]
- Greer, N.D. Voriconazole: The newest triazole antifungal agent. Bayl. Univ. Med. Cent. Proc. 2003, 16, 241–248. [Google Scholar] [CrossRef]
- Richard, M.; Ibata-Ombetta, S.; Dromer, F.; Bordon-Pallier, F.; Jouault, T.; Gaillardin, C. Complete glycosylphosphatidylinositol anchors are required in Candida albicans for full morphogenesis, virulence and resistance to macrophages. Mol. Microbiol. 2002, 44, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Taira, T. Structures and antifungal activity of plant chitinases. J. Appl. Glyosc. 2010, 57, 167–176. [Google Scholar] [CrossRef]
- Shimizu, K.; Keller, N.P. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 2001, 157, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Liu, Y.; Wang, S.; Wu, L.; Xie, R.; Lan, H.; Fasoyin, O.E.; Wang, Y.; Wang, S. Cyclase-associated protein cap with multiple domains contributes to mycotoxin biosynthesis and fungal virulence in Aspergillus flavus. J. Agric. Food Chem. 2019, 67, 4200–4213. [Google Scholar] [CrossRef] [PubMed]
- Weidner, G.; d’Enfert, C.; Koch, A.; Mol, P.C.; Brakhage, A.A. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5’-monophosphate decarboxylase. Curr. Genet. 1998, 33, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Skory, C.D.; Horng, J.S.; Pestka, J.J.; Linz, J.E. Transformation of Aspergillus parasiticus with a homologous gene (pyrG) involved in pyrimidine biosynthesis. Appl. Environ. Microbiol. 1990, 56, 3315–3320. [Google Scholar] [CrossRef]
- Lan, H.; Sun, R.; Fan, K.; Yang, K.; Zhang, F.; Nie, X.; Wang, X.; Zhuang, Z.; Wang, S. The Aspergillus flavus histone acetyltransferase AflGcnE regulates morphogenesis, aflatoxin biosynthesis, and pathogenicity. Front. Microbiol. 2016, 7, 1324. [Google Scholar] [CrossRef]
- Yang, K.; Liang, L.; Ran, F.; Liu, Y.; Li, Z.; Lan, H.; Gao, P.; Zhuang, Z.; Zhang, F.; Nie, X.; et al. The DmtA methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Sci. Rep. 2016, 6, 23259. [Google Scholar] [CrossRef]
- Zhang, F.; Xu, G.; Geng, L.; Lu, X.; Yang, K.; Yuan, J.; Nie, X.; Zhuang, Z.; Wang, S. The stress response regulator AflSkn7 influences morphological development, stress response, and pathogenicity in the fungus Aspergillus flavus. Toxins 2016, 8, 202. [Google Scholar] [CrossRef]
- Chang, P.K.; Scharfenstein, L.L.; Wei, Q.; Bhatnagar, D. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J. Microbiol. Methods 2010, 81, 240–246. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Wen, M.; Li, G.; Wang, S. Chitin Deacetylase Homologous Gene cda Contributes to Development and Aflatoxin Synthesis in Aspergillus flavus. Toxins 2024, 16, 217. https://doi.org/10.3390/toxins16050217
Zhang X, Wen M, Li G, Wang S. Chitin Deacetylase Homologous Gene cda Contributes to Development and Aflatoxin Synthesis in Aspergillus flavus. Toxins. 2024; 16(5):217. https://doi.org/10.3390/toxins16050217
Chicago/Turabian StyleZhang, Xin, Meifang Wen, Guoqi Li, and Shihua Wang. 2024. "Chitin Deacetylase Homologous Gene cda Contributes to Development and Aflatoxin Synthesis in Aspergillus flavus" Toxins 16, no. 5: 217. https://doi.org/10.3390/toxins16050217



