Recent Advances in Microfluidic-Based Extracellular Vesicle Analysis
Abstract
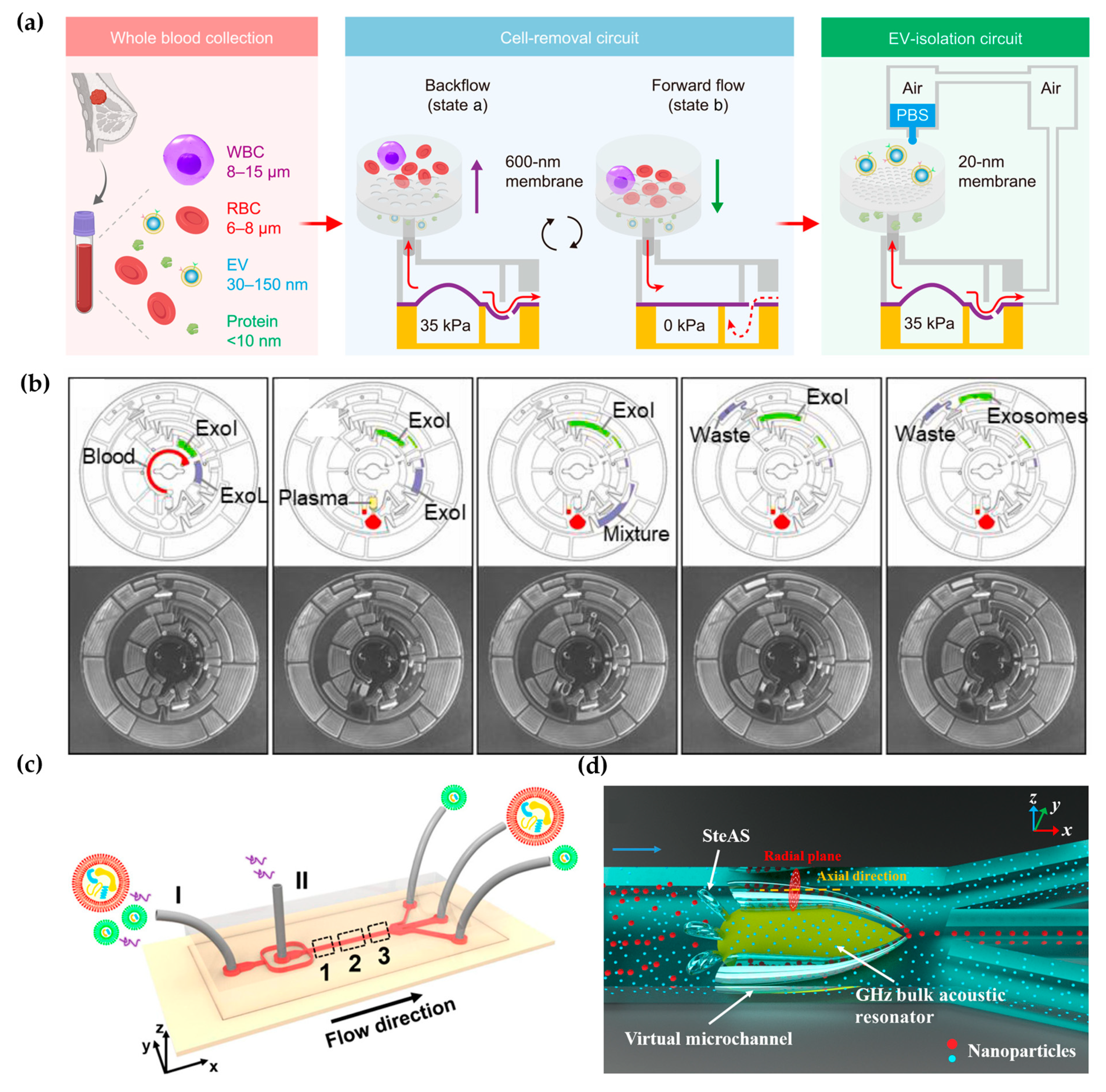
:1. Introduction
2. Microfluidic-Based EV Isolation Strategies
2.1. Label-Free Microfluidic Isolation
2.2. Affinity-Based EV Isolation
3. Microfluidic-Based EV Detection
3.1. Fluorescent Detection
3.2. Visualization Detection
3.3. Electrochemical Detection
3.4. Surface-Enhanced Raman Spectroscopy (SERS)
4. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Zhu, L.; Tian, W.; Yuan, L.; Chi, C.; Wang, Y.; Xiao, Q.; Zheng, M.; Yang, C.; Song, Y. Aptamer-Based Extracellular Vesicle Isolation, Analysis and Therapeutics. Interdiscip. Med. 2023, 1, e20220019. [Google Scholar] [CrossRef]
- Robbins, P.; Morelli, A. Regulation of Immune Responses by Extracellular Vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V. The Biology Function and Biomedical Applications of Exosomes. Science 2020, 367, 6977. [Google Scholar] [CrossRef]
- Möller, A.; Lobb, R. The Evolving Translational Potential of Small Extracellular Vesicles in Cancer. Nat. Rev. Can. 2020, 20, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Bio. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Peng, C.; Yi, J.; Zhang, D.; Xiang, X.; Peng, X.; Su, B.; Liu, B.; Shen, Y.; Qiao, L. Highly Efficient Exosome Purification from Human Plasma by Tangential Flow Filtration Based Microfluidic Chip. Sens. Actuators B Chem. 2021, 333, 129563. [Google Scholar] [CrossRef]
- Li, P.; Chen, J.; Chen, Y.; Song, S.; Huang, X.; Yang, Y.; Li, Y.; Tong, Y.; Xie, Y.; Li, J.; et al. Construction of Exosome Sorl1 Detection Platform Based on 3d Porous Microfluidic Chip and Its Application in Early Diagnosis of Colorectal Cancer. Small 2023, 19, e2207381. [Google Scholar] [CrossRef] [PubMed]
- Coumans, F.; Brisson, A.; Buzas, E.; Dignat-George, F.; Drees, E.; El-Andaloussi, S.; Emanueli, C.; Gasecka, A.; Hendrix, A.; Hill, A.; et al. Methodological Guidelines to Study Extracellular Vesicles. Circ. Res. 2017, 120, 1632–1648. [Google Scholar] [CrossRef]
- Whitesides, G. The Origins and the Future of Microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, S.; Dunlay, C.; Simeone, D.; Nagrath, S. Microfluidic Device (Exochip) for on-Chip Isolation, Quantification and Characterization of Circulating Exosomes. Lab Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef] [PubMed]
- Royo, F.; Théry, C.; Falcón-Pérez, J.M.; Nieuwland, R.; Witwer, K.W. Methods for Separation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey Performed by the Isev Rigor and Standardization Subcommittee. Cells 2020, 9, 1955. [Google Scholar] [CrossRef]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Nolte-t’ Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of Sample Collection, Isolation and Analysis Methods in Extracellular Vesicle Research. J. Extracell. Vesicles. 2013, 2, e1700716. [Google Scholar] [CrossRef]
- Kalra, H.; Adda, C.G.; Liem, M.; Ang, C.S.; Mechler, A.; Simpson, R.J.; Hulett, M.D.; Mathivanan, S. Comparative Proteomics Evaluation of Plasma Exosome Isolation Techniques and Assessment of the Stability of Exosomes in Normal Human Blood Plasma. Proteomics 2013, 13, 3354–3364. [Google Scholar] [CrossRef]
- Alvarez, M.; Khosroheidari, M.; Kanchi Ravi, R.; DiStefano, J. Comparison of Protein, Microrna, and Mrna Yields Using Different Methods of Urinary Exosome Isolation for the Discovery of Kidney Disease Biomarkers. Kidney Int. 2012, 82, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Nordin, J.Z.; Lee, Y.; Vader, P.; Mäger, I.; Johansson, H.J.; Heusermann, W.; Wiklander, O.P.; Hällbrink, M.; Seow, Y.; Bultema, J.J.; et al. Ultrafiltration with Size-Exclusion Liquid Chromatography for High Yield Isolation of Extracellular Vesicles Preserving Intact Biophysical and Functional Properties. Nanomedicine 2015, 11, 879–883. [Google Scholar] [CrossRef] [PubMed]
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef] [PubMed]
- Poudineh, M.; Aldridge, P.; Ahmed, S.; Green, B.; Kermanshah, L.; Nguyen, V.; Tu, C.; Mohamadi, R.; Nam, R.; Hansen, A.; et al. Tracking the Dynamics of Circulating Tumour Cell Phenotypes Using Nanoparticle-Mediated Magnetic Ranking. Nat. Nanotechnol. 2017, 12, 274–281. [Google Scholar] [CrossRef]
- Ibsen, S.; Wright, J.; Lewis, J.; Kim, S.; Ko, S.; Ong, J.; Manouchehri, S.; Vyas, A.; Akers, J.; Chen, C.; et al. Rapid Isolation and Detection of Exosomes and Associated Biomarkers from Plasma. ACS Nano 2017, 11, 6641–6651. [Google Scholar] [CrossRef]
- Psaltis, D.; Quake, S.; Yang, C. Developing Optofluidic Technology through the Fusion of Microfluidics and Optics. Nature 2006, 442, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Li-Guo, L.; Meng-Qi, K.; Sherry, Z.; Ye-Feng, S.; Ping, W.; Tao, Y.; Fatih, I.; Winston Patrick, K.; Lan-Juan, L.; Utkan, D.; et al. An Integrated Double-Filtration Microfluidic Device for Isolation, Enrichment and Quantification of Urinary Extracellular Vesicles for Detection of Bladder Cancer. Sci. Rep. 2017, 7, 46224. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, C.; Cheng, Y.; Li, Y.; Deng, J.; Bai, L.; Qin, L.; Mei, H.; Zeng, M.; Tian, F.; et al. Cascaded Microfluidic Circuits for Pulsatile Filtration of Extracellular Vesicles from Whole Blood for Early Cancer Diagnosis. Sci. Adv. 2023, 9, eade2819. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, H.; Fu, J.; Wu, X.; Liang, X.Y.; Liu, X.Y.; Wu, X.; Cao, L.L.; Xu, Z.Y.; Dong, M. Microfluidic-Based Exosome Isolation and Highly Sensitive Aptamer Exosome Membrane Protein Detection for Lung Cancer Diagnosis. Biosens. Bioelectron. 2022, 214, 114487. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, A.; Xiaobao, C.; Bogdan, M.; Daniel, V.L.; Mahmut Kamil, A.; Stavros, S.; Andrew, J.d. Oscillatory Viscoelastic Microfluidics for Efficient Focusing and Separation of Nanoscale Species. ACS Nano 2019, 14, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Guo, J.; Tian, F.; Yang, N.; Yan, F.; Ding, Y.; Wei, J.; Hu, G.; Nie, G.; Sun, J. Field-Free Isolation of Exosomes from Extracellular Vesicles by Microfluidic Viscoelastic Flows. ACS Nano 2017, 11, 6968–6976. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, L.; Jin, K.; He, M.; Wei, W.; Chen, X.; Yang, Q.; Wang, Y.; Pang, W.; Ren, X.; et al. Self-Adaptive Virtual Microchannel for Continuous Enrichment and Separation of Nanoparticles. Sci. Adv. 2022, 8, eabn8440. [Google Scholar] [CrossRef] [PubMed]
- Salafi, T.; Zhang, Y.; Zhang, Y. A Review on Deterministic Lateral Displacement for Particle Separation and Detection. Nano-Micro Lett. 2019, 11, 77–109. [Google Scholar] [CrossRef] [PubMed]
- Kerwin Kwek, Z.; Thoriq, S.; Swati, S.; Yong, Z. Fluorescent Label-Free Quantitative Detection of Nano-Sized Bioparticles Using a Pillar Array. Nat. Commun. 2018, 9, 1254. [Google Scholar] [CrossRef]
- Benjamin, H.W.; Joshua, T.S.; Stacey, M.G.; Chao, W.; Markus, B.; Robert, L.B.; Robert, H.A.; Gustavo, S.; Yann, A. Nanoscale Lateral Displacement Arrays for the Separation of Exosomes and Colloids Down to 20 Nm. Nat. Nanotechnol. 2016, 11, 936–940. [Google Scholar] [CrossRef]
- Joshua, T.S.; Benjamin, H.W.; Navneet, D.; Mehmet, E.A.; Kayla, L.; Kamlesh, K.Y.; Rachel, W.; Michael, A.P.; Jyotica, V.P.; Elizabeth, A.D.; et al. Integrated Nanoscale Deterministic Lateral Displacement Arrays for Separation of Extracellular Vesicles from Clinically-Relevant Volumes of Biological Samples. Lab Chip 2018, 18, 3913–3925. [Google Scholar] [CrossRef] [PubMed]
- Yuya, H.; Taisuke, S.; Takao, Y.; Noritada, K.; Yoshinobu, B. Micro- and Nanopillar Chips for Continuous Separation of Extracellular Vesicles. Anal. Chem. 2019, 91, 6514–6521. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Im, H.; Castro, C.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef] [PubMed]
- Boriachek, K.; Masud, M.; Palma, C.; Phan, H.; Yamauchi, Y.; Hossain, M.; Nguyen, N.; Salomon, C.; Shiddiky, M. Avoiding Pre-Isolation Step in Exosome Analysis: Direct Isolation and Sensitive Detection of Exosomes Using Gold-Loaded Nanoporous Ferric Oxide Nanozymes. Anal. Chem. 2019, 91, 3827–3834. [Google Scholar] [CrossRef] [PubMed]
- Evgen, M.; Crystal Jing Ying, T.; Mari, P.; Pia, S.; Marja-Liisa, R. Fast Isolation of Highly Specific Population of Platelet-Derived Extracellular Vesicles from Blood Plasma by Affinity Monolithic Column, Immobilized with Anti-Human Cd61 Antibody. Anal. Chim. Acta 2019, 1091, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.K.; Whiteside, T.L. Immunoaffinity-Based Isolation of Melanoma Cell-Derived and T Cell-Derived Exosomes from Plasma of Melanoma Patients. Methods Mol. Biol. 2021, 2265, 305–321. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, J.; Lau, S.; Akbarinejad, A.; Bryant, D.T.; Chamley, L.W.; Pilkington, L.I.; Barker, D.; Williams, D.E.; Evans, C.W.; Travas-Sejdic, J. Electrochemical Approach for Specific Capture and Rapid Release of Nanoscale Placental Extracellular Vesicles Using Aptamer-Modified Conducting Terpolymer-Coated Carbon Cloth. ACS Appl. Nano Mater. 2023, 6, 3981–3989. [Google Scholar] [CrossRef]
- Huang, W.; Yu, Y.; Yang, C.; Zhang, X.; Shang, L.; Zu, Y.; Shi, K. Aptamer Decorated Magnetic Graphene Oxide Nanoparticles for Effective Capture of Exosomes. Chem. Eng. J. 2022, 431, 133849. [Google Scholar] [CrossRef]
- Qiannan, Y.; Linlin, Z.; Zhimin, C.; Mingfeng, G.; Qian, M.; Li, Y.; Wen-Fei, D. Hierarchical Au Nanoarrays Functionalized 2d Ti(2)Ct(X) Mxene Membranes for the Detection of Exosomes Isolated from Human Lung Carcinoma Cells. Biosens. Bioelectron. 2022, 216, 14647. [Google Scholar] [CrossRef]
- Chen, H.; Bian, F.; Guo, J.; Zhao, Y. Aptamer-Functionalized Barcodes in Herringbone Microfluidics for Multiple Detection of Exosomes. Small Methods 2022, 6, e2200236. [Google Scholar] [CrossRef]
- Niu, Q.; Gao, J.; Zhao, K.; Chen, X.; Lin, X.; Huang, C.; An, Y.; Xiao, X.; Wu, Q.; Cui, L.; et al. Fluid Nanoporous Microinterface Enables Multiscale-Enhanced Affinity Interaction for Tumor-Derived Extracellular Vesicle Detection. Proc. Natl. Acad. Sci. USA 2022, 119, e2213236119. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.K.; Enderle, D.; Noerholm, M.; Breakefield, X.O.; Skog, J.K. Exosome-Based Liquid Biopsies in Cancer: Opportunities and Challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef]
- Niu, Q.; Shu, Y.; Chen, Y.; Huang, Z.; Yao, Z.; Chen, X.; Lin, F.; Feng, J.; Huang, C.; Wang, H.; et al. A Fluid Multivalent Magnetic Interface for High-Performance Isolation and Proteomic Profiling of Tumor-Derived Extracellular Vesicles. Angew. Chem. Int. Ed. 2023, 62, 202215337. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, Q.; Huang, Y.; Wang, W.; Lu, Y.; Kang, S.; Yang, C.; Song, Y. Reliable Detection of Extracellular Pd-L1 by DNA Computation-Mediated Microfluidics. Anal. Chem. 2023, 95, 9373–9379. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Duong, B.; Ahmed, S.; Dhavarasa, P.; Wang, Z.; Labib, M.; Flynn, C.; Xu, J.; Zhang, Y.; Wang, H.; et al. A Magneto-Activated Nanoscale Cytometry Platform for Molecular Profiling of Small Extracellular Vesicles. Nat. Commun. 2023, 14, 5576. [Google Scholar] [CrossRef] [PubMed]
- Yinzhu, L.; Bingqian, L.; Weizhi, L.; Jialu, Z.; Lin, Z.; Chaoyong, Y.; Yanling, S. Isolation of Pd-L1 Extracellular Vesicle Subpopulations Using DNA Computation Mediated Microfluidic Tandem Separation. Small Methods 2023, 7, e2300516. [Google Scholar] [CrossRef] [PubMed]
- Mun, B.; Kim, R.; Jeong, H.; Kang, B.; Kim, J.; Son, H.Y.; Lim, J.; Rho, H.W.; Lim, E.K.; Haam, S. An Immuno-Magnetophoresis-Based Microfluidic Chip to Isolate and Detect Her2-Positive Cancer-Derived Exosomes Via Multiple Separation. Biosens. Bioelectron. 2023, 239, 115592. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (Misev2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the Misev2014 Guidelines. J. Extracell. Vesicles. 2018, 7, 535750. [Google Scholar] [CrossRef]
- Cheng, S.; Li, Y.; Yan, H.; Wen, Y.; Zhou, X.; Friedman, L.; Zeng, Y. Advances in Microfluidic Extracellular Vesicle Analysis for Cancer Diagnostics. Lab Chip 2021, 21, 3219–3243. [Google Scholar] [CrossRef]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A Protocol for Exosome Isolation and Characterization: Evaluation of Ultracentrifugation, Density-Gradient Separation, and Immunoaffinity Capture Methods. Methods Mol. Biol. 2015, 1295, 179–209. [Google Scholar] [CrossRef]
- Takov, K.; Yellon, D.M.; Davidson, S.M. Comparison of Small Extracellular Vesicles Isolated from Plasma by Ultracentrifugation or Size-Exclusion Chromatography: Yield, Purity and Functional Potential. J. Extracell. Vesicles. 2019, 8, 1560809. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Du, D.; Wang, X.; Liu, D.; Xu, W.; Luo, Y.; Lin, Y. Recent Advances in Biosensors for Detecting Cancer-Derived Exosomes. Trends. Biotech. 2019, 37, 1236–1254. [Google Scholar] [CrossRef] [PubMed]
- Dudani, J.; Gossett, D.; Tse, H.; Lamm, R.; Kulkarni, R.; Carlo, D. Rapid Inertial Solution Exchange for Enrichment and Flow Cytometric Detection of Microvesicles. Biomicrofluidics 2015, 9, 014112. [Google Scholar] [CrossRef] [PubMed]
- Hisey, C.; Dorayappan, K.; Cohn, D.; Selvendiran, K.; Hansford, D. Microfluidic Affinity Separation Chip for Selective Capture and Release of Label-Free Ovarian Cancer Exosomes. Lab Chip 2018, 18, 3144–3153. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Ye, L.; Jian, X.; Yang, D.; Zhang, H.; Tong, Z.; Wu, Z.; Shi, N.; Han, Y.; Mao, H. Integrated Microfluidic System for Isolating Exosome and Analyzing Protein Marker Pd-L1. Biosens. Bioelectron. 2022, 204, 113879. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xu, X.; Li, B.; Situ, B.; Pan, W.; Hu, Y.; An, T.; Yao, S.; Zheng, L. Single-Exosome-Counting Immunoassays for Cancer Diagnostics. Nano Lett. 2018, 18, 4226–4232. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.Q.; Zhao, S.; Li, J.H.; Cheng, Y.C.; Liu, C.; Liu, Z.; Li, L.L.; Tian, F.; Dai, B.; Sun, J.S. One-Step Thermophoretic and Gate Operation on Extracellular Vesicles Improves Diagnosis of Prostate Cancer. Angew. Chem. Int. Ed. 2022, 61, e202207037. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, S.H.; Hu, H.J.; Cheng, Y.C.; Zou, K.X.; Song, J.; Deng, J.Q.; Li, L.L.; Zhang, X.B.; Ke, G.L.; et al. Selective in Situ Analysis of Mature Micrornas in Extracellular Vesicles Using a DNA Cage-Based Thermophoretic Assay. Angew. Chem. Int. Ed. 2023, 62, 202303121. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.H.; Qi, D.Y.; Li, L.M.; Su, H.R.; Li, X.J.; Luo, Y.; Sun, B.; Zhang, F.Y.; Lin, B.C.; Liu, T.J.; et al. Multiplexed Profiling of Single-Cell Extracellular Vesicles Secretion. Proc. Natl. Acad. Sci. USA 2019, 116, 5979–5984. [Google Scholar] [CrossRef]
- Zhang, P.; Zhou, X.; He, M.; Shang, Y.; Tetlow, A.; Godwin, A.; Zeng, Y. Ultrasensitive Detection of Circulating Exosomes with a 3d-Nanopatterned Microfluidic Chip. Nat. Biomed. Eng. 2019, 3, 438–451. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, X.; Gardashova, G.; Yang, Y.; Zhang, Y.; Xu, L.; Zeng, Y. Molecular and Functional Extracellular Vesicle Analysis Using Nanopatterned Microchips Monitors Tumor Progression and Metastasis. Sci. Transl. Med. 2020, 12, eaaz2878. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Yuanfeng, X.; Xinyu, W.; Haoting, L.; Mengjiao, H.; Bingqian, L.; Yanling, S.; Chaoyong, Y. Coupling Aptamer-Based Protein Tagging with Metabolic Glycan Labeling for in Situ Visualization and Biological Function Study of Exosomal Protein-Specific Glycosylation. Angew. Chem. Int. Ed. 2021, 60, 18111–18115. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhao, J.; Tian, F.; Cai, L.; Zhang, W.; Feng, Q.; Chang, J.; Wan, F.; Yang, Y.; Dai, B.; et al. Low-Cost Thermophoretic Profiling of Extracellular-Vesicle Surface Proteins for the Early Detection and Classification of Cancers. Nat. Biomed. Eng. 2019, 3, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Zhang, S.H.; Liu, C.; Han, Z.W.; Liu, Y.; Deng, J.Q.; Li, Y.K.; Wu, X.; Cai, L.L.; Qin, L.L.; et al. Protein Analysis of Extracellular Vesicles to Monitor and Predict Therapeutic Response in Metastatic Breast Cancer. Nat. Commun. 2021, 12, 2536. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Deng, J.; Han, Z.; Liu, C.; Tian, F.; Xu, R.; Han, D.; Zhang, S.; Sun, J. Molecular Identification of Tumor-Derived Extracellular Vesicles Using Thermophoresis-Mediated DNA Computation. J. Am. Chem. Soc. 2021, 143, 1290–1295. [Google Scholar] [CrossRef] [PubMed]
- Thind, A.; Wilson, C. Exosomal Mirnas as Cancer Biomarkers and Therapeutic Targets. J. Extracell. Vesicles 2016, 5, 31292. [Google Scholar] [CrossRef] [PubMed]
- Huilin, S.; Jaehoon, C.; Kyungheon, L.; Leonora, B.; Changwook, M.; Bob, S.C.; Fred, H.H.; Xandra, O.B.; Hakho, L.; Ralph, W. Chip-Based Analysis of Exosomal Mrna Mediating Drug Resistance in Glioblastoma. Nat. Commun. 2015, 6, 6999. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, C.; Li, Y.; Ma, Y.; Deng, J.; Li, L.; Sun, J. Thermophoretic Detection of Exosomal Micrornas by Nanoflares. J. Am. Chem. Soc. 2020, 142, 4996–5001. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Wan, F.; Deng, J.; Zhao, J.; Li, Y.; Yang, Y.; Jiang, Q.; Ding, B.; Liu, C.; Dai, B.; et al. Ultrasensitive Detection of Mrna in Extracellular Vesicles Using DNA Tetrahedron-Based Thermophoretic Assay. Nano Today 2021, 38, 101203. [Google Scholar] [CrossRef]
- Crescitelli, R.; Lässer, C.; Lötvall, J. Isolation and Characterization of Extracellular Vesicle Subpopulations from Tissues. Nat. Protoc. 2021, 16, 1548–1580. [Google Scholar] [CrossRef]
- Deng, J.; Ji, Y.; Zhu, F.; Liu, L.; Li, L.; Bai, X.; Li, H.; Liu, X.; Luo, Y.; Lin, B.; et al. Mapping Secretome-Mediated Interaction between Paired Neuron–Macrophage Single Cells. Proc. Natl. Acad. Sci. USA 2022, 119, e2200944119. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cheng, S.; Cao, P.; Qiu, Q.; Chen, Y.; Xie, M.; Xu, Y.; Huang, W. Detection of Exosomes by Zno Nanowires Coated Three-Dimensional Scaffold Chip Device. Biosens. Bioelectron. 2018, 122, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Di, H.; Mi, Z.; Sun, Y.; Liu, X.; Liu, X.; Li, A.; Jiang, Y.; Gao, H.; Rong, P.; Liu, D. Nanozyme-Assisted Sensitive Profiling of Exosomal Proteins for Rapid Cancer Diagnosis. Theranostics 2020, 10, 9303–9314. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Shi, M.; Liu, Y.; Wan, S.; Cui, C.; Zhang, L.; Tan, W. Aptamer/Aunp Biosensor for Colorimetric Profiling of Exosomal Proteins. Angew. Chem. Int. Ed. 2017, 56, 11916–11920. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Hemphill, M.; Gabrieli, D.; Wu, L.; Yelleswarapu, V.; Lawrence, G.; Pennycooke, W.; Singh, A.; Meaney, D.; Issadore, D. Smartphone-Enabled Optofluidic Exosome Diagnostic for Concussion Recovery. Sci. Rep. 2016, 6, 31215. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Park, J.S.; Huang, C.-H.; Jo, A.; Cook, K.; Wang, R.; Lin, H.-Y.; Van Deun, J.; Li, H.; Min, J.; et al. An Integrated Magneto-Electrochemical Device for the Rapid Profiling of Tumour Extracellular Vesicles from Blood Plasma. Nat. Biomed. Eng. 2021, 5, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Takanori, A.; Kei, K.; Masashi, K.; Nobuyoshi, K.; Takahiro, O.; Takanori, I. On-Chip Immunoelectrophoresis of Extracellular Vesicles Released from Human Breast Cancer Cells. PLoS ONE 2015, 10, e0123603. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Kobayashi, M.; Hanamura, N.; Akagi, T.; Kosaka, N.; Ochiya, T.; Ichiki, T. Electrokinetic Evaluation of Individual Exosomes by on-Chip Microcapillary Electrophoresis with Laser Dark-Field Microscopy. Jpn. J. Appl. Phys. 2013, 52, 06GK10. [Google Scholar] [CrossRef]
- Akagi, T.; Kato, K.; Hanamura, N.; Kobayashi, M.; Ichiki, T. Evaluation of Desialylation Effect on Zeta Potential of Extracellular Vesicles Secreted from Human Prostate Cancer Cells by on-Chip Microcapillary Electrophoresis. Jpn. J. Appl. Phys. 2014, 53, 06JL01. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, W.; Sun, M.; Feng, B.; Shen, H.; Zhu, J.; Chen, X.; Yu, S. A Filter-Electrochemical Microfluidic Chip for Multiple Surface Protein Analysis of Exosomes to Detect and Classify Breast Cancer. Biosens. Bioelectron. 2023, 239, 115590. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Z.; Li, G.; Jing, A. Room-Temperature Self-Healing Conductive Elastomers for Modular Assembly as a Microfluidic Electrochemical Biosensing Platform for the Detection of Colorectal Cancer Exosomes. Micromachines 2023, 14, 617. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liao, C.; Zuo, P.; Liu, Z.; Ye, B. Magnetic-Based Microfluidic Device for on-Chip Isolation and Detection of Tumor-Derived Exosomes. Anal. Chem. 2018, 90, 13451–13458. [Google Scholar] [CrossRef]
- Huang, R.; He, L.; Xia, Y.; Xu, H.; Liu, C.; Xie, H.; Wang, S.; Peng, L.; Liu, Y.; Liu, Y.; et al. A Sensitive Aptasensor Based on a Hemin/G-Quadruplex-Assisted Signal Amplification Strategy for Electrochemical Detection of Gastric Cancer Exosomes. Small 2019, 15, e1900735. [Google Scholar] [CrossRef]
- Zhang, W.; Tian, Z.; Yang, S.; Rich, J.; Zhao, S.; Klingeborn, M.; Huang, P.; Li, Z.; Stout, A.; Murphy, Q.; et al. Electrochemical Micro-Aptasensors for Exosome Detection Based on Hybridization Chain Reaction Amplification. Microsyst. Nanoeng. 2021, 7, 63. [Google Scholar] [CrossRef]
- Chen, M.; Lin, S.; Zhou, C.; Cui, D.; Haick, H.; Tang, N. From Conventional to Microfluidic: Progress in Extracellular Vesicle Separation and Individual Characterization. Adv. Healthc. Mater. 2023, 12, e2202437. [Google Scholar] [CrossRef]
- Panneerselvam, R.; Sadat, H.; Höhn, E.; Das, A.; Noothalapati, H.; Belder, D. Microfluidics and Surface-Enhanced Raman Spectroscopy, a Win-Win Combination? Lab Chip 2022, 22, 665–682. [Google Scholar] [CrossRef]
- Yue, S.; Fang, J.; Xu, Z. Advances in Droplet Microfluidics for Sers and Raman Analysis. Biosens. Bioelectron. 2022, 198, 113822. [Google Scholar] [CrossRef]
- Jalali, M.; Isaac Hosseini, I.; AbdelFatah, T.; Montermini, L.; Wachsmann Hogiu, S.; Rak, J.; Mahshid, S. Plasmonic Nanobowtiefluidic Device for Sensitive Detection of Glioma Extracellular Vesicles by Raman Spectrometry. Lab Chip 2021, 21, 855–866. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Q.; Shi, H.; Tang, K.; Qiao, L.; Yu, G.; Ding, C.; Yu, S. Microfluidic Raman Biochip Detection of Exosomes: A Promising Tool for Prostate Cancer Diagnosis. Lab Chip 2020, 20, 4632–4637. [Google Scholar] [CrossRef]
- Wang, J.; Wuethrich, A.; Sina, A.; Lane, R.; Lin, L.; Wang, Y.; Cebon, J.; Behren, A.; Trau, M. Tracking Extracellular Vesicle Phenotypic Changes Enables Treatment Monitoring in Melanoma. Sci. Adv. 2020, 6, eaax3223. [Google Scholar] [CrossRef]
- Han, Z.; Peng, X.; Yang, Y.; Yi, J.; Zhao, D.; Bao, Q.; Long, S.; Yu, S.; Xu, X.; Liu, B.; et al. Integrated Microfluidic-Sers for Exosome Biomarker Profiling and Osteosarcoma Diagnosis. Biosens. Bioelectron. 2022, 217, 114709. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Vaidyanathan, R.; Shiddiky, M.; Trau, M. Enabling Rapid and Specific Surface-Enhanced Raman Scattering Immunoassay Using Nanoscaled Surface Shear Forces. ACS Nano 2015, 9, 6354–6362. [Google Scholar] [CrossRef] [PubMed]




| Aspect | Traditional Techniques | Microfluidic-Based Methodologies |
|---|---|---|
| Effectiveness | Varied, depending on method (e.g., ultracentrifugation, precipitation) | High, with precise control over fluid manipulation and surface interactions |
| Efficiency | Time-consuming, labor-intensive | Rapid, automated processes with minimal sample and reagent consumption |
| Practicality | Limited scalability, manual operation | Scalable, integrated systems suitable for high-throughput analysis |
| Specificity | May lack specificity, leading to contamination and low yield | Enhanced specificity, with tailored devices for selective EV capture |
| Reproducibility | Variable due to manual handling and batch-to-batch variability | Improved reproducibility with standardized protocols and automated workflows |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Zheng, M.; Xiao, Q.; Wang, H.; Chi, C.; Lin, T.; Wang, Y.; Yi, X.; Zhu, L. Recent Advances in Microfluidic-Based Extracellular Vesicle Analysis. Micromachines 2024, 15, 630. https://doi.org/10.3390/mi15050630
Chen J, Zheng M, Xiao Q, Wang H, Chi C, Lin T, Wang Y, Yi X, Zhu L. Recent Advances in Microfluidic-Based Extracellular Vesicle Analysis. Micromachines. 2024; 15(5):630. https://doi.org/10.3390/mi15050630
Chicago/Turabian StyleChen, Jiming, Meiyu Zheng, Qiaoling Xiao, Hui Wang, Caixing Chi, Tahui Lin, Yulin Wang, Xue Yi, and Lin Zhu. 2024. "Recent Advances in Microfluidic-Based Extracellular Vesicle Analysis" Micromachines 15, no. 5: 630. https://doi.org/10.3390/mi15050630





