Floxed Il1rl2 Locus with mCherry Reporter Element Reveals Distinct Expression Patterns of the IL-36 Receptor in Barrier Tissues
Abstract
:1. Introduction
2. Materials and Methods
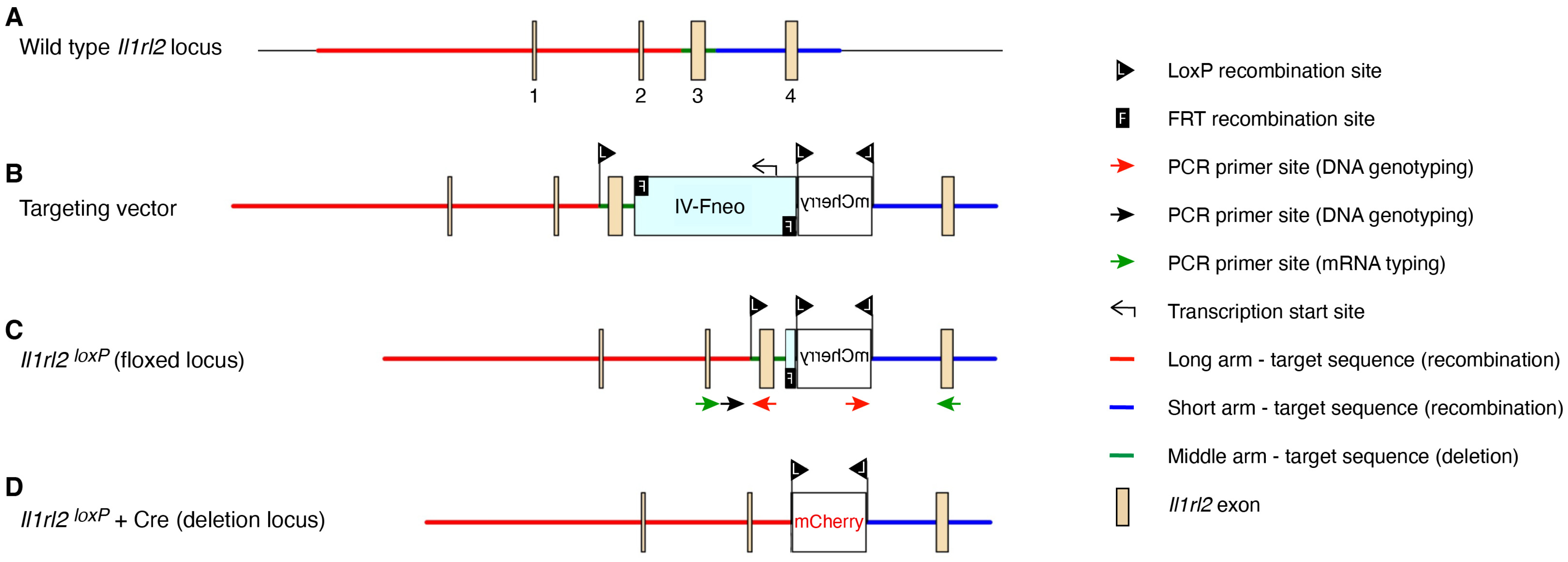
2.1. Generation of Floxed Il1rl2 Locus
2.2. Generation of Cre-Expressing Strains and Genotyping
2.3. Tissue Collection
2.4. RNA Isolation and Analyses
2.5. Herpes Simplex Virus-1 (HSV-1) Infections
2.6. Immunohistochemistry and Immunofluorescence
3. Results
3.1. Il1rl2 Targeting Strategy
3.2. Functional Validation of Floxed Locus
3.3. Functional Validation of mCherry Reporter in Skin
3.4. Application of mCherry Reporter to Identify IL-1RL2 Expressing Cells in Barrier Tissues
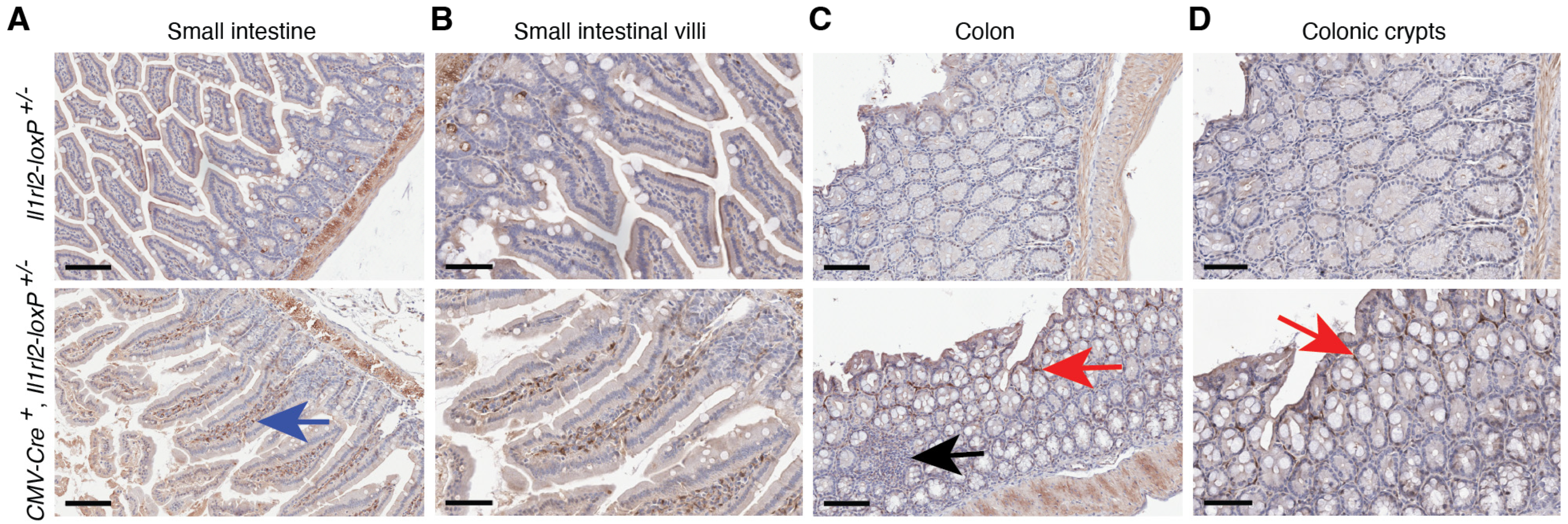
3.4.1. Small Intestine and Colon
3.4.2. Lungs and Trachea
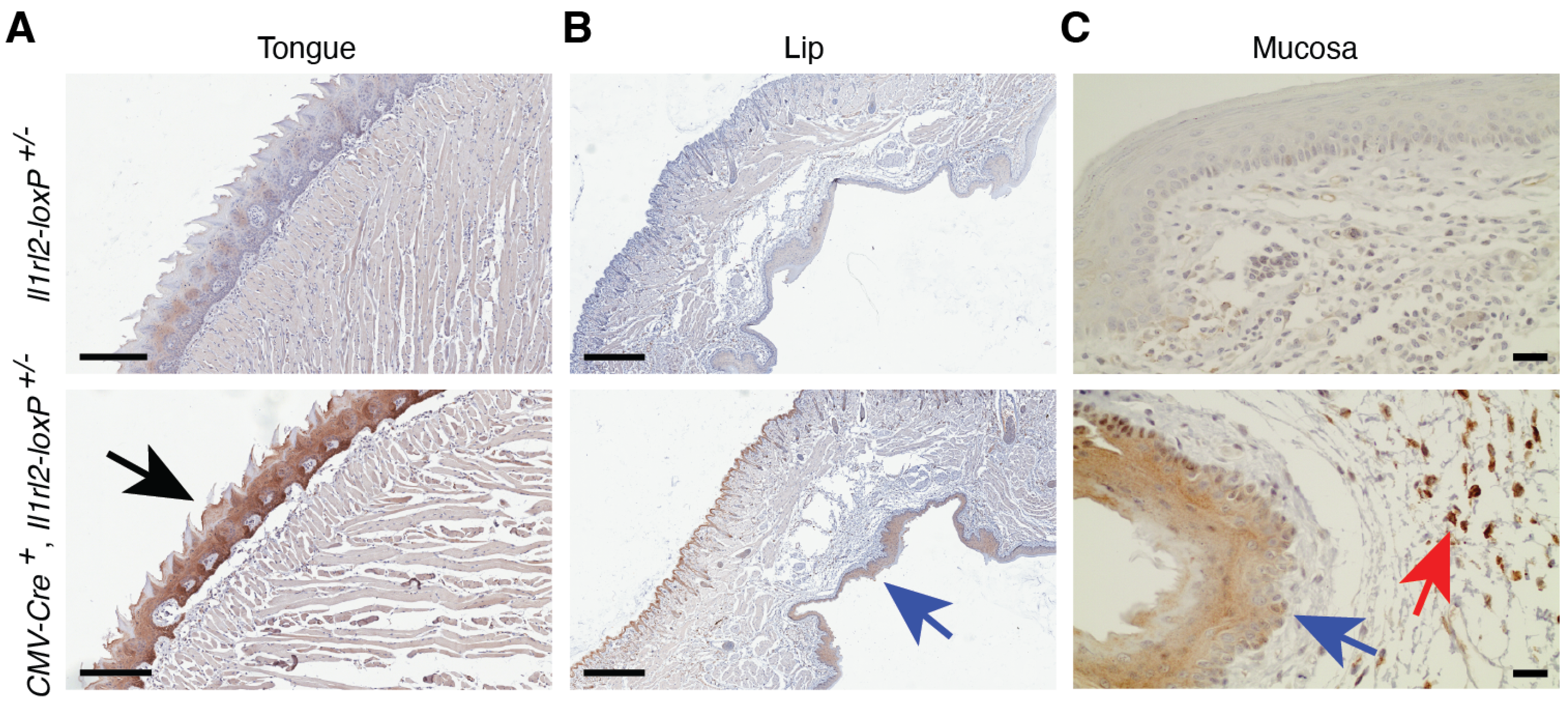
3.4.3. Oral Mucosa and Tongue
3.4.4. Vagina and Uterus
3.4.5. Esophagus and Stomach
3.4.6. Urinary Tract
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Blair, H.A. Spesolimab: First Approval. Drugs 2022, 82, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Paulsboe, S.; Wetter, J.; Salte, K.; Kannan, A.; Mathew, S.; Horowitz, A.; Gerstein, C.; Namovic, M.; Todorović, V.; et al. IL-36 receptor antagonistic antibodies inhibit inflammatory responses in preclinical models of psoriasiform dermatitis. Exp. Dermatol. 2019, 28, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, R.; Raymond, E.L.; Mennerich, D.; Woska, J.R., Jr.; Caviness, G.; Grimaldi, C.; Ahlberg, J.; Perez, R.; Roberts, S.; Yang, D.; et al. Generation and functional characterization of anti-human and anti-mouse IL-36R antagonist monoclonal antibodies. MAbs 2017, 9, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Iznardo, H.; Puig, L. Exploring the Role of IL-36 Cytokines as a New Target in Psoriatic Disease. Int. J. Mol. Sci. 2021, 22, 4344. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, S.L.; Bai, A.; Bailey, D.; Ichikawa, K.; Zielinski, J.; Karp, R.; Apte, A.; Arnold, K.; Zacharek, S.J.; Iliou, M.S.; et al. Durable anticancer immunity from intratumoral administration of IL-23, IL-36γ, and OX40L mRNAs. Sci. Transl. Med. 2019, 11, eaat9143. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.E. Interleukin-36 cytokines may overcome microbial immune evasion strategies that inhibit interleukin-1 family signaling. Sci. Signal. 2017, 10, eaan3589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Jiang, M.; Huang, M.; Yang, J.; Liu, Q.; Zhao, Z.; Bai, Y.; He, T.; Zhang, D.; Zhang, M. Prognostic value of Interleukin-36s in cancers: A systematic review and meta-analysis. Cytokine 2023, 172, 156397. [Google Scholar] [CrossRef] [PubMed]
- Tsurutani, N.; Mittal, P.; St Rose, M.-C.; Ngoi, S.M.; Svedova, J.; Menoret, A.; Treadway, F.B.; Laubenbacher, R.; Suárez-Ramírez, J.E.; Cauley, L.S.; et al. Costimulation endows immunotherapeutic CD8 T cells with IL-36 responsiveness during aerobic glycolysis. J. Immunol. 2016, 196, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Gao, D.S.; Shoush, J.; Lu, B. The IL-1 family in tumorigenesis and antitumor immunity. Semin. Cancer Biol. 2022, 86, 280–295. [Google Scholar] [CrossRef]
- Okorie, C.L.; Nayudu, K.; Nambudiri, V.E. Cutaneous findings and treatments in deficiency of interleukin-36 receptor antagonist (DITRA): A review of the literature. Exp. Dermatol. 2023, 33, e14934. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Alam, M.A.; Ansari, A.W.; Jochebeth, A.; Leo, R.; Al-Abdulla, M.N.; Al-Khawaga, S.; AlHammadi, A.; Al-Malki, A.; Al Naama, K.; et al. Emerging Role of the IL-36/IL-36R Axis in Multiple Inflammatory Skin Diseases. J. Investig. Dermatol. 2024, 144, 206–224. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.S.; Sun, X.; Wong, C.K. The Role of New IL-1 Family Members (IL-36 and IL-38) in Atopic Dermatitis, Allergic Asthma, and Allergic Rhinitis. Curr. Allergy Asthma Rep. 2020, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Hao, Y.; Li, W.; Yang, W.; Gao, P. IL-36 Cytokines: Their Roles in Asthma and Potential as a Therapeutic. Front. Immunol. 2022, 13, 921275. [Google Scholar] [CrossRef] [PubMed]
- Byrne, J.; Baker, K.; Houston, A.; Brint, E. IL-36 cytokines in inflammatory and malignant diseases: Not the new kid on the block anymore. Cell. Mol. Life Sci. 2021, 78, 6215–6227. [Google Scholar] [CrossRef] [PubMed]
- Neurath, M.F. Targeting cytokines in inflammatory bowel disease. Sci. Transl. Med. 2022, 14, eabq4473. [Google Scholar] [CrossRef] [PubMed]
- Foster, A.M.; Baliwag, J.; Chen, C.S.; Guzman, A.M.; Stoll, S.W.; Gudjonsson, J.E.; Ward, N.L.; Johnston, A. IL-36 promotes myeloid cell infiltration, activation, and inflammatory activity in skin. J. Immunol. 2014, 192, 6053–6061. [Google Scholar] [CrossRef] [PubMed]
- Mutamba, S.; Allison, A.; Mahida, Y.; Barrow, P.; Foster, N. Expression of IL-1Rrp2 by human myelomonocytic cells is unique to DCs and facilitates DC maturation by IL-1F8 and IL-1F9. Eur. J. Immunol. 2012, 42, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Vigne, S.; Palmer, G.; Lamacchia, C.; Martin, P.; Talabot-Ayer, D.; Rodriguez, E.; Ronchi, F.; Sallusto, F.; Dinh, H.; Sims, J.E.; et al. IL-36R ligands are potent regulators of dendritic and T cells. Blood 2011, 18, 5813–5823. [Google Scholar] [CrossRef] [PubMed]
- Vigne, S.; Palmer, G.; Martin, P.; Lamacchia, C.; Strebel, D.; Rodriguez, E.; Olleros, M.L.; Vesin, D.; Garcia, I.; Ronchi, F.; et al. IL-36 signaling amplifies Th1 responses by enhancing proliferation and Th1 polarization of naive CD4+ T cells. Blood 2012, 120, 3478–3487. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Archer, N.K.; Dillen, C.A.; Wang, Y.; Ashbaugh, A.G.; Ortines, R.V.; Kao, T.; Lee, S.K.; Cai, S.S.; Miller, R.J.; et al. Staphylococcus aureus Epicutaneous Exposure Drives Skin Inflammation via IL-36-Mediated T Cell Responses. Cell Host Microbe 2017, 22, 653–666. [Google Scholar] [CrossRef]
- Patrick, G.J.; Liu, H.; Alphonse, M.P.; Dikeman, D.A.; Youn, C.; Otterson, J.C.; Wang, Y.; Ravipati, A.; Mazhar, M.; Denny, G.; et al. Epicutaneous Staphylococcus aureus induces IL-36 to enhance IgE production and ensuing allergic disease. J. Clin. Investig. 2021, 131, e143334. [Google Scholar] [CrossRef] [PubMed]
- Yarmolinsky, M.; Hoess, R. The Legacy of Nat Sternberg: The Genesis of Cre-lox Technology. Annu. Rev. Virol. 2015, 2, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.E.; Hoess, K.; Mitchell, L.E.; Whitehead, A.S. Loss of function polymorphisms in NAT1 protect against spina bifida. Hum. Genet. 2006, 120, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Milora, K.A.; Uppalapati, S.R.; Sanmiguel, J.C.; Zou, W.; Jensen, L.E. Interleukin-36β provides protection against HSV-1 infection, but does not modulate initiation of adaptive immune responses. Sci. Rep. 2017, 7, 5799. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Gamero, A.M.; Jensen, L.E. IL-36 promotes anti-viral immunity by boosting sensitivity to IFN-alpha/beta in IRF1 dependent and independent manners. Nat. Commun. 2019, 10, 4700. [Google Scholar] [CrossRef] [PubMed]
- Milora, K.A.; Fu, H.; Dubaz, O.; Jensen, L.E. Unprocessed interleukin-36α regulates psoriasis-like skin inflammation in co-operation with interleukin-1. J. Investig. Dermatol. 2015, 135, 2992–3000. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Herranz, M.; Lian, L.-H.; Hooper, K.M.; Milora, K.A.; Jensen, L.E. IL-1R1 signaling facilitates Munro’s microabscess formation in psoriasiform imiquimod-induced skin inflammation. J. Investig. Dermatol. 2013, 133, 1541–1549. [Google Scholar] [CrossRef]
- Milora, K.A.; Miller, S.L.; Sanmiguel, J.C.; Jensen, L.E. Interleukin-1α released from HSV-1 infected keratinocytes acts as a functional alarmin in the skin. Nat. Commun. 2014, 5, 5230. [Google Scholar] [CrossRef] [PubMed]
- Towne, J.E.; Garka, K.E.; Renshaw, B.R.; Virca, G.D.; Sims, J.E. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J. Biol. Chem. 2004, 279, 13677–13688. [Google Scholar] [CrossRef] [PubMed]
- Tortola, L.; Rosenwald, E.; Abel, B.; Blumberg, H.; Schäfer, M.; Coyle, A.J.; Renauld, J.-C.; Werner, S.; Kisielow, J.; Kopf, M. Psoriasiform dermatitis is driven by IL-36-mediated DC-keratinocyte crosstalk. J. Clin. Investig. 2012, 122, 3965–3976. [Google Scholar] [CrossRef] [PubMed]
- Medina-Contreras, O.; Harusato, A.; Nishio, H.; Flannigan, K.L.; Ngo, V.; Leoni, G.; Neumann, P.-A.; Geem, D.; Lili, L.N.; Ramadas, R.A.; et al. Cutting edge: IL-36 receptor promotes resolution of intestinal damage. J. Immunol. 2016, 196, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Scheibe, K.; Backert, I.; Wirtz, S.; Hueber, A.; Schett, G.; Vieth, M.; Probst, H.C.; Bopp, T.; Neurath, M.F.; Neufert, C. IL-36R signalling activates intestinal epithelial cells and fibroblasts and promotes mucosal healing in vivo. Gut 2016, 66, 823–838. [Google Scholar] [CrossRef] [PubMed]
- Boutet, M.A.; Bart, G.; Penhoat, M.; Amiaud, J.; Brulin, B.; Charrier, C.; Morel, F.; Lecron, J.C.; Rolli-Derkinderen, M.; Bourreille, A.; et al. Distinct expression of interleukin (IL)-36α, β and γ, their antagonist IL-36Ra and IL-38 in psoriasis, rheumatoid arthritis and Crohn’s disease. Clin. Exp. Immunol. 2016, 184, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Nishida, A.; Hidaka, K.; Kanda, T.; Imaeda, H.; Shioya, M.; Inatomi, O.; Bamba, S.; Kitoh, K.; Sugimoto, M.; Andoh, A. Increased Expression of Interleukin-36, a Member of the Interleukin-1 Cytokine Family, in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2016, 22, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Nishida, A.; Takahashi, K.; Hidaka, K.; Imaeda, H.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Andoh, A. Interleukin(IL)-36α and IL-36γ Induce Proinflammatory Mediators from Human Colonic Subepithelial Myofibroblasts. Front. Med. 2015, 2, 69. [Google Scholar] [CrossRef]
- Sun, H.; Tan, J.; Chen, H.; Wu, N.; Su, B. Immune niches orchestrated by intestinal mesenchymal stromal cells lining the crypt-villus. Front. Immunol. 2022, 13, 1057932. [Google Scholar] [CrossRef] [PubMed]
- Ramadas, R.A.; Ewart, S.L.; Iwakura, Y.; Medoff, B.D.; LeVine, A.M. IL-36alpha exerts pro-inflammatory effects in the lungs of mice. PLoS ONE 2012, 7, e45784. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, T.; Newstead, M.W.; Zeng, X.; Kunkel, S.L.; Kaku, M.; Standiford, T.J. IL-36 receptor deletion attenuates lung injury and decreases mortality in murine influenza pneumonia. Mucosal Immunol. 2017, 10, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, T.; Newstead, M.W.; Zeng, X.; Nanjo, Y.; Peters-Golden, M.; Kaku, M.; Standiford, T.J. Interleukin-36γ and IL-36 receptor signaling mediate impaired host immunity and lung injury in cytotoxic Pseudomonas aeruginosa pulmonary infection: Role of prostaglandin E2. PLoS Pathog. 2017, 13, e1006737. [Google Scholar] [CrossRef] [PubMed]
- Kovach, M.A.; Singer, B.; Martinez-Colon, G.; Newstead, M.W.; Zeng, X.; Mancuso, P.; Moore, T.A.; Kunkel, S.L.; Peters-Golden, M.; Moore, B.B.; et al. IL-36γ is a crucial proximal component of protective type-1-mediated lung mucosal immunity in Gram-positive and -negative bacterial pneumonia. Mucosal Immunol. 2017, 10, 1320. [Google Scholar] [CrossRef] [PubMed]
- Wein, A.N.; Dunbar, P.R.; McMaster, S.R.; Li, Z.-R.T.; Denning, T.L.; Kohlmeier, J.E. IL-36γ protects against severe influenza infection by promoting lung alveolar macrophage survival and limiting viral replication. J. Immunol. 2018, 201, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Nanjo, Y.; Newstead, M.W.; Aoyagi, T.; Zeng, X.; Takahashi, K.; Yu, F.S.; Tateda, K.; Standiford, T.J. Overlapping Roles for Interleukin-36 Cytokines in Protective Host Defense against Murine Legionella pneumophila Pneumonia. Infect. Immun. 2019, 87, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Koss, C.K.; Wohnhaas, C.T.; Baker, J.R.; Tilp, C.; Przibilla, M.; Lerner, C.; Frey, S.; Keck, M.; Williams, C.M.M.; Peter, D.; et al. IL36 is a critical upstream amplifier of neutrophilic lung inflammation in mice. Commun. Biol. 2021, 4, 172. [Google Scholar] [CrossRef] [PubMed]
- Heath, J.E.; Scholz, G.M.; Veith, P.D.; Reynolds, E.C. IL-36γ regulates mediators of tissue homeostasis in epithelial cells. Cytokine 2019, 119, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Scholz, G.M.; Heath, J.E.; Walsh, K.A.; Reynolds, E.C. MEK-ERK signaling diametrically controls the stimulation of IL-23p19 and EBI3 expression in epithelial cells by IL-36γ. Immunol. Cell Biol. 2018, 96, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Huynh, J.; Scholz, G.M.; Aw, J.; Kwa, M.Q.; Achuthan, A.; Hamilton, J.A.; Reynolds, E.C. IRF6 regulates the expression of IL-36γ by human oral epithelial cells in response to Porphyromonas gingivalis. J. Immunol. 2016, 196, 2230–2238. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Shao, R.R.; Zhang, S.; Tan, Z.W.; Guo, Y.T.; He, Y. The mechanism on Prevotella melaninogenica promoting the inflammatory progression of oral lichen planus. Clin. Exp. Immunol. 2022, 209, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.H.; Zafar, H.; Ponde, N.O.; Hepworth, O.W.; Sihra, D.; Aggor, F.E.Y.; Ainscough, J.S.; Ho, J.; Richardson, J.P.; Coleman, B.M.; et al. IL-36 and IL-1/IL-17 Drive Immunity to Oral Candidiasis via Parallel Mechanisms. J. Immunol. 2018, 201, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Meng, H.; Mao, Y.; Zhong, L.; Pan, W.; Chen, Q. IL-36 Regulates Neutrophil Chemotaxis and Bone Loss at the Oral Barrier. J. Dent. Res. 2024, 103, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Vo, P.T.; Choi, S.S.; Park, H.R.; Lee, A.; Jeong, S.H.; Choi, Y. Gene signatures associated with barrier dysfunction and infection in oral lichen planus identified by analysis of transcriptomic data. PLoS ONE 2021, 16, e0257356. [Google Scholar] [CrossRef] [PubMed]
- Cloitre, A.; Halgand, B.; Sourice, S.; Caillon, J.; Huck, O.; Bugueno, I.M.; Batool, F.; Guicheux, J.; Geoffroy, V.; Lesclous, P. IL-36γ is a pivotal inflammatory player in periodontitis-associated bone loss. Sci. Rep. 2019, 9, 19257. [Google Scholar] [CrossRef] [PubMed]
- Babaloo, A.R.; Shirmohammadi, A.; Sandoghchian, S.; Kamalzadeh, A.; Ghasemi, S. Evaluation of the effect of IL-36γ expression on chronic periodontitis by enhancing the MAPK and TLR4 signaling pathways: A basic research. J. Dent. Res. Dent. Clin. Dent. Prospects 2018, 12, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Winkle, S.M.; Throop, A.L.; Herbst-Kralovetz, M.M. IL-36γ augments host defense and immune responses in human female reproductive tract epithelial cells. Front. Microbiol. 2016, 7, 955. [Google Scholar] [CrossRef] [PubMed]
- Gardner, J.K.; Herbst-Kralovetz, M.M. IL-36γ induces a transient HSV-2 resistant environment that protects against genital disease and pathogenesis. Cytokine 2018, 111, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Gardner, J.K.; Swaims-Kohlmeier, A.; Herbst-Kralovetz, M.M. IL-36γ is a key regulator of neutrophil infiltration in the vaginal microenvironment and limits neuroinvasion in genital HSV-2 infection. J. Immunol. 2019, 203, 2655–2664. [Google Scholar] [CrossRef] [PubMed]
- Stanfield, B.A.; Rider, P.J.F.; Caskey, J.; Del Piero, F.; Kousoulas, K.G. Intramuscular vaccination of guinea pigs with the live-attenuated human herpes simplex vaccine VC2 stimulates a transcriptional profile of vaginal Th17 and regulatory Tr1 responses. Vaccine 2018, 36, 2842–2849. [Google Scholar] [CrossRef] [PubMed]
- Gardner, J.K.; Łaniewski, P.; Knight, A.; Haddad, L.B.; Swaims-Kohlmeier, A.; Herbst-Kralovetz, M.M. Interleukin-36γ Is Elevated in Cervicovaginal Epithelial Cells in Women with Bacterial Vaginosis and In Vitro After Infection with Microbes Associated With Bacterial Vaginosis. J. Infect. Dis. 2020, 221, 983–988. [Google Scholar] [CrossRef] [PubMed]
- Birse, K.D.; Romas, L.M.; Guthrie, B.L.; Nilsson, P.; Bosire, R.; Kiarie, J.; Farquhar, C.; Broliden, K.; Burgener, A.D. Genital Injury Signatures and Microbiome Alterations Associated with Depot Medroxyprogesterone Acetate Usage and Intravaginal Drying Practices. J. Infect. Dis. 2017, 215, 590–598. [Google Scholar] [CrossRef]
- Łaniewski, P.; Barnes, D.; Goulder, A.; Cui, H.; Roe, D.J.; Chase, D.M.; Herbst-Kralovetz, M.M. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci. Rep. 2018, 8, 7593. [Google Scholar] [CrossRef] [PubMed]
- Lovenberg, T.W.; Crowe, P.D.; Liu, C.; Chalmers, D.T.; Liu, X.J.; Liaw, C.; Clevenger, W.; Oltersdorf, T.; De Souza, E.B.; Maki, R.A. Cloning of a cDNA encoding a novel interleukin-1 receptor related protein (IL 1R-rp2). J. Neuroimmunol. 1996, 70, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Debets, R.; Timans, J.C.; Homey, B.; Zurawski, S.; Sana, T.R.; Lo, S.; Wagner, J.; Edwards, G.; Clifford, T.; Menon, S.; et al. Two novel IL-1 family members, IL-1 delta and IL-1 epsilon, function as an antagonist and agonist of NF-kappa B activation through the orphan IL-1 receptor-related protein 2. J. Immunol. 2001, 167, 1440–1446. [Google Scholar] [CrossRef] [PubMed]
- Koop, K.; Enderle, K.; Hillmann, M.; Ruspeckhofer, L.; Vieth, M.; Sturm, G.; Trajanoski, Z.; Kühl, A.A.; Atreya, R.; Leppkes, M.; et al. Interleukin 36 receptor-inducible matrix metalloproteinase 13 mediates intestinal fibrosis. Front. Immunol. 2023, 14, 1163198. [Google Scholar] [CrossRef] [PubMed]
- Ngo, V.L.; Abo, H.; Maxim, E.; Harusato, A.; Geem, D.; Medina-Contreras, O.; Merlin, D.; Gewirtz, A.T.; Nusrat, A.; Denning, T.L. A cytokine network involving IL-36γ, IL-23, and IL-22 promotes antimicrobial defense and recovery from intestinal barrier damage. Proc. Natl. Acad. Sci. USA 2018, 115, E5076–E5085. [Google Scholar] [CrossRef] [PubMed]
- Harusato, A.; Abo, H.; Ngo, V.L.; Yi, S.W.; Mitsutake, K.; Osuka, S.; Kohlmeier, J.E.; Li, J.D.; Gewirtz, A.T.; Nusrat, A.; et al. IL-36γ signaling controls the induced regulatory T cell-Th9 cell balance via NFκB activation and STAT transcription factors. Mucosal. Immunol. 2017, 10, 1455–1467. [Google Scholar] [CrossRef] [PubMed]
- Ngo, V.L.; Abo, H.; Kuczma, M.; Szurek, E.; Moore, N.; Medina-Contreras, O.; Nusrat, A.; Merlin, D.; Gewirtz, A.T.; Ignatowicz, L.; et al. IL-36R signaling integrates innate and adaptive immune-mediated protection against enteropathogenic bacteria. Proc. Natl. Acad. Sci. USA 2020, 117, 27540–27548. [Google Scholar] [CrossRef] [PubMed]
- Tomuschat, C.; O’Donnell, A.M.; Coyle, D.; Puri, P. Altered expression of IL36γ and IL36 receptor (IL1RL2) in the colon of patients with Hirschsprung’s disease. Pediatr. Surg. Int. 2017, 33, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Ohno, M.; Imai, T.; Chatani, M.; Nishida, A.; Inatomi, O.; Kawahara, M.; Hoshino, T.; Andoh, A. The anti-inflammatory and protective role of interleukin-38 in inflammatory bowel disease. J. Clin. Biochem. Nutr. 2022, 70, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Narros-Fernández, P.; Chomanahalli Basavarajappa, S.; Walsh, P.T. Interleukin-1 family cytokines at the crossroads of microbiome regulation in barrier health and disease. FEBS J. 2023, 291, 1849–1869. [Google Scholar] [CrossRef] [PubMed]
- Capone, K.A.; Dowd, S.E.; Stamatas, G.N.; Nikolovski, J. Diversity of the human skin microbiome early in life. J. Investig. Dermatol. 2011, 131, 2026–2032. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Program, N.C.S.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Findley, K.; Oh, J.; Yang, J.; Conlan, S.; Deming, C.; Meyer, J.A.; Schoenfeld, D.; Nomicos, E.; Park, M.; Kong, H.H.; et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013, 498, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Hannigan, G.D.; Meisel, J.S.; Tyldsley, A.S.; Zheng, Q.; Hodkinson, B.P.; SanMiguel, A.J.; Minot, S.; Bushman, F.D.; Grice, E.A. The human skin double-stranded DNA virome: Topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. mBio 2015, 6, e01578-15. [Google Scholar] [CrossRef] [PubMed]







| Forward Primers | Reverse Primers |
| LOX1: AGGGAAGCTGTCTTTAGAACCAAGC | SDL2: CACACGTATCTGGGGAAGGAAAGG |
| PNDEL1: TCCCAAGTCTCCCTCTCCAT | PNDEL2: CCATCTGTTGTTTGCCCCTC |
| CHRRYSC1: CACCCTTGGTCACCTTCAGCTTGG | A2: ACCTTCAAGGACCTGTGTCATTCC |
| Il1rl2E2: TTGCTCTTCTGTGGGGTGTTT | Il1rl2E4: GGTAGCAGTTGTGGGCATTC |
| Cre: GCGGTCTGGCAGTAAAAACTATC | Cre: GTGAAACAGCATTGCTGTCACTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tongmuang, N.; Cai, K.Q.; An, J.; Novy, M.; Jensen, L.E. Floxed Il1rl2 Locus with mCherry Reporter Element Reveals Distinct Expression Patterns of the IL-36 Receptor in Barrier Tissues. Cells 2024, 13, 787. https://doi.org/10.3390/cells13090787
Tongmuang N, Cai KQ, An J, Novy M, Jensen LE. Floxed Il1rl2 Locus with mCherry Reporter Element Reveals Distinct Expression Patterns of the IL-36 Receptor in Barrier Tissues. Cells. 2024; 13(9):787. https://doi.org/10.3390/cells13090787
Chicago/Turabian StyleTongmuang, Nopprarat, Kathy Q. Cai, Jiahui An, Mariah Novy, and Liselotte E. Jensen. 2024. "Floxed Il1rl2 Locus with mCherry Reporter Element Reveals Distinct Expression Patterns of the IL-36 Receptor in Barrier Tissues" Cells 13, no. 9: 787. https://doi.org/10.3390/cells13090787





