Machine Learning for Short-Term Mortality in Acute Decompensation of Liver Cirrhosis: Better than MELD Score
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Data
2.3. Development of ML Model
2.4. Statistical Analysis
3. Results
3.1. Baseline Characteristics
3.2. MELD-Na and MELD 3.0 for Predicting 28- and 90-Day Mortality in Whole Sample
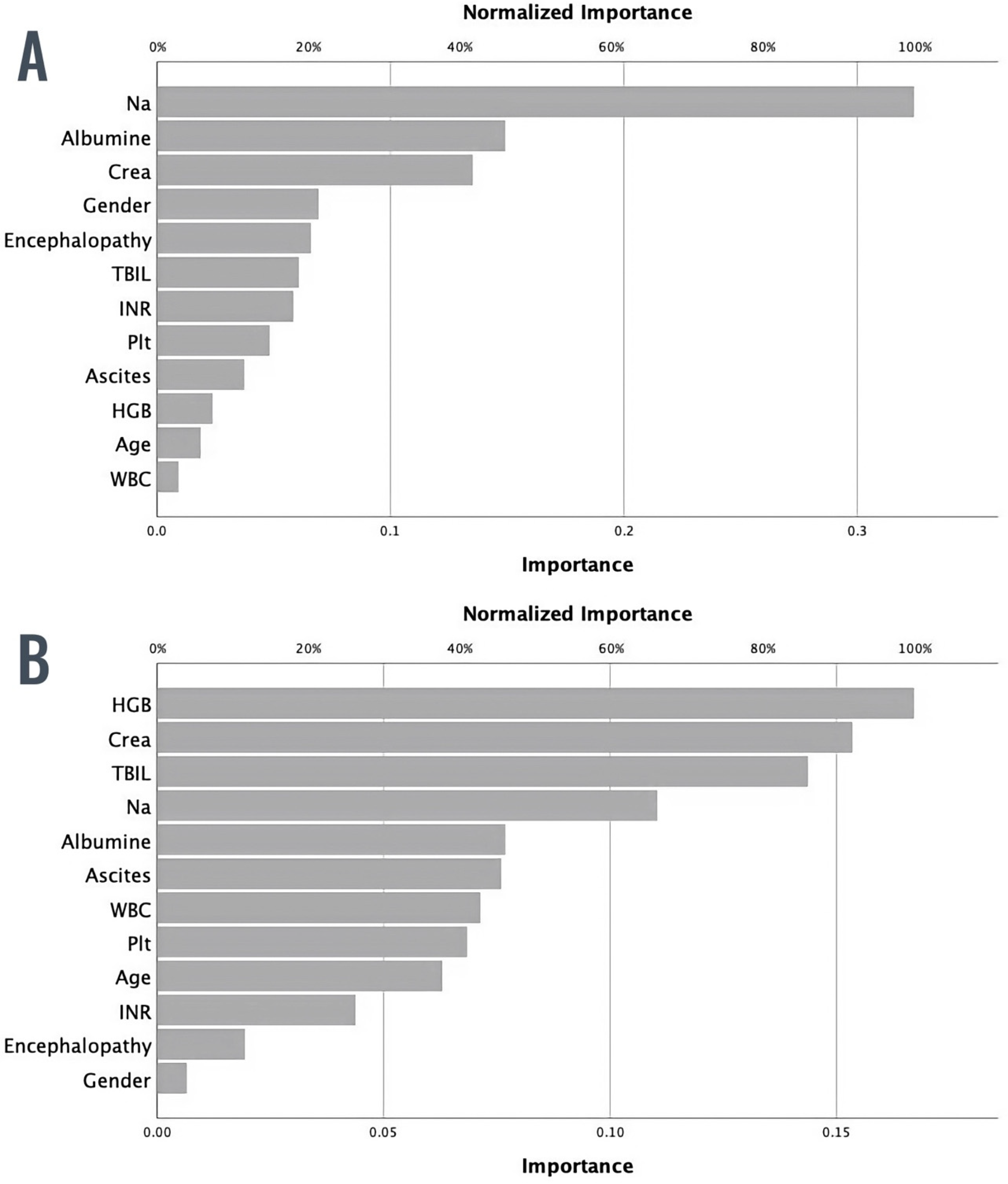
3.3. ML Model for Predicting 28-Day Mortality
3.4. ML Model for Predicting 90-Day Mortality
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.-B.; Chen, M.-K. Epidemiology of Liver Cirrhosis and Associated Complications: Current Knowledge and Future Directions. World J. Gastroenterol. 2022, 28, 5910–5930. [Google Scholar] [CrossRef] [PubMed]
- Sepanlou, S.G.; Safiri, S.; Bisignano, C.; Ikuta, K.S.; Merat, S.; Saberifiroozi, M.; Poustchi, H.; Tsoi, D.; Colombara, D.V.; Abdoli, A.; et al. The Global, Regional, and National Burden of Cirrhosis by Cause in 195 Countries and Territories, 1990–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Zhai, M.; Long, J.; Gong, Y.; Ren, C.; Zhang, D.; Lin, X.; Liu, S. The Burden of Liver Cirrhosis in Mortality: Results from the Global Burden of Disease Study. Front. Public Health 2022, 10, 909455. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, G.; Garcia-Tsao, G.; Pagliaro, L. Natural History and Prognostic Indicators of Survival in Cirrhosis: A Systematic Review of 118 Studies. J. Hepatol. 2006, 44, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Jochmans, I.; van Rosmalen, M.; Pirenne, J.; Samuel, U. Adult Liver Allocation in Eurotransplant. Transplantation 2017, 101, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Latt, N.L.; Niazi, M.; Pyrsopoulos, N.T. Liver Transplant Allocation Policies and Outcomes in United States: A Comprehensive Review. World J. Methodol. 2022, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.R.; Mannalithara, A.; Heimbach, J.K.; Kamath, P.S.; Asrani, S.K.; Biggins, S.W.; Wood, N.L.; Gentry, S.E.; Kwong, A.J. MELD 3.0: The Model for End-Stage Liver Disease Updated for the Modern Era. Gastroenterology 2021, 161, 1887–1895.e4. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.R.; Biggins, S.W.; Kremers, W.K.; Wiesner, R.H.; Kamath, P.S.; Benson, J.T.; Edwards, E.; Therneau, T.M. Hyponatremia and Mortality among Patients on the Liver-Transplant Waiting List. N. Engl. J. Med. 2008, 359, 1018–1026. [Google Scholar] [CrossRef] [PubMed]
- Biggins, S.W.; Rodriguez, H.J.; Bacchetti, P.; Bass, N.M.; Roberts, J.P.; Terrault, N.A. Serum Sodium Predicts Mortality in Patients Listed for Liver Transplantation. Hepatology 2005, 41, 32–39. [Google Scholar] [CrossRef]
- Pugh, R.N.; Murray-Lyon, I.M.; Dawson, J.L.; Pietroni, M.C.; Williams, R. Transection of the Oesophagus for Bleeding Oesophageal Varices. Br. J. Surg. 1973, 60, 646–649. [Google Scholar] [CrossRef]
- Nagai, S.; Chau, L.C.; Schilke, R.E.; Safwan, M.; Rizzari, M.; Collins, K.; Yoshida, A.; Abouljoud, M.S.; Moonka, D. Effects of Allocating Livers for Transplantation Based on Model for End-Stage Liver Disease–Sodium Scores on Patient Outcomes. Gastroenterology 2018, 155, 1451–1462.e3. [Google Scholar] [CrossRef]
- Ruf, A.; Dirchwolf, M.; Freeman, R.B. From Child-Pugh to MELD Score and beyond: Taking a Walk down Memory Lane. Ann. Hepatol. 2022, 27, 100535. [Google Scholar] [CrossRef]
- Dziodzio, T.; Öllinger, R.; Schöning, W.; Rothkäppel, A.; Nikolov, R.; Juraszek, A.; Ritschl, P.V.; Stockmann, M.; Pratschke, J.; Jara, M. Validation of a New Prognostic Model to Predict Short and Medium-Term Survival in Patients with Liver Cirrhosis. BMC Gastroenterol. 2020, 20, 265. [Google Scholar] [CrossRef]
- Shehab, M.; Abualigah, L.; Shambour, Q.; Abu-Hashem, M.A.; Shambour, M.K.Y.; Alsalibi, A.I.; Gandomi, A.H. Machine Learning in Medical Applications: A Review of State-of-the-Art Methods. Comput. Biol. Med. 2022, 145, 105458. [Google Scholar] [CrossRef]
- Quazi, S. Artificial Intelligence and Machine Learning in Precision and Genomic Medicine. Med. Oncol. 2022, 39, 120. [Google Scholar] [CrossRef]
- Cucchetti, A.; Vivarelli, M.; Heaton, N.D.; Phillips, S.; Piscaglia, F.; Bolondi, L.; La Barba, G.; Foxton, M.R.; Rela, M.; O’Grady, J.; et al. Artificial Neural Network Is Superior to MELD in Predicting Mortality of Patients with End-Stage Liver Disease. Gut 2007, 56, 253–258. [Google Scholar] [CrossRef]
- Mai, R.-Y.; Zeng, J.; Mo, Y.-S.; Liang, R.; Lin, Y.; Wu, S.-S.; Piao, X.-M.; Gao, X.; Wu, G.-B.; Li, L.-Q.; et al. Artificial Neural Network Model for Liver Cirrhosis Diagnosis in Patients with Hepatitis B Virus-Related Hepatocellular Carcinoma. Ther. Clin. Risk Manag. 2020, 16, 639–649. [Google Scholar] [CrossRef]
- Hou, Y.; Zhang, Q.; Gao, F.; Mao, D.; Li, J.; Gong, Z.; Luo, X.; Chen, G.; Li, Y.; Yang, Z.; et al. Artificial Neural Network-Based Models Used for Predicting 28- and 90-Day Mortality of Patients with Hepatitis B-Associated Acute-on-Chronic Liver Failure. BMC Gastroenterol. 2020, 20, 75. [Google Scholar] [CrossRef]
- Ghoshal, U.C.; Das, A. Models for Prediction of Mortality from Cirrhosis with Special Reference to Artificial Neural Network: A Critical Review. Hepatol. Int. 2008, 2, 31–38. [Google Scholar] [CrossRef]
- Guo, A.; Mazumder, N.R.; Ladner, D.P.; Foraker, R.E. Predicting Mortality among Patients with Liver Cirrhosis in Electronic Health Records with Machine Learning. PLoS ONE 2021, 16, e0256428. [Google Scholar] [CrossRef]
- Banerjee, R.; Das, A.; Ghoshal, U.C.; Sinha, M. Predicting Mortality in Patients with Cirrhosis of Liver with Application of Neural Network Technology. J. Gastroenterol. Hepatol. 2003, 18, 1054–1060. [Google Scholar] [CrossRef]
- Tonon, M.; Moreau, R. Using Machine Learning for Predicting Outcomes in ACLF. Liver Int. 2022, 42, 2354–2355. [Google Scholar] [CrossRef]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine Learning in Medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine Learning in Medicine: A Practical Introduction. BMC Med. Res. Methodol. 2019, 19, 64. [Google Scholar] [CrossRef]
- D’Amico, G.; Bernardi, M.; Angeli, P. Towards a New Definition of Decompensated Cirrhosis. J. Hepatol. 2022, 76, 202–207. [Google Scholar] [CrossRef]
- Weissenborn, K. Hepatic Encephalopathy: Definition, Clinical Grading and Diagnostic Principles. Drugs 2019, 79, 5–9. [Google Scholar] [CrossRef]
- Rojas, R. The Backpropagation Algorithm. In Neural Networks; Springer: Berlin/Heidelberg, Germany, 1996; pp. 149–182. ISBN 978-3-540-60505-8. [Google Scholar]
- Alqahtani, S.A.; Gurakar, A.; Tamim, H.; Schiano, T.D.; Bonder, A.; Fricker, Z.; Kazimi, M.; Eckhoff, D.E.; Curry, M.P.; Saberi, B. Regional and National Trends of Adult Living Donor Liver Transplantation in the United States Over the Last Two Decades. J. Clin. Transl. Hepatol. 2022, 10, 814–824. [Google Scholar] [CrossRef]
- Penny, W.; Frost, D. Neural Networks in Clinical Medicine. Med. Decis. Mak. 1996, 16, 386–398. [Google Scholar] [CrossRef]
- Haglin, J.M.; Jimenez, G.; Eltorai, A.E.M. Artificial Neural Networks in Medicine. Health Technol. 2019, 9, 1–6. [Google Scholar] [CrossRef]
- Yu, C.; Li, Y.; Yin, M.; Gao, J.; Xi, L.; Lin, J.; Liu, L.; Zhang, H.; Wu, A.; Xu, C.; et al. Automated Machine Learning in Predicting 30-Day Mortality in Patients with Non-Cholestatic Cirrhosis. J. Pers. Med. 2022, 12, 1930. [Google Scholar] [CrossRef]
- Kanwal, F.; Taylor, T.J.; Kramer, J.R.; Cao, Y.; Smith, D.; Gifford, A.L.; El-Serag, H.B.; Naik, A.D.; Asch, S.M. Development, Validation, and Evaluation of a Simple Machine Learning Model to Predict Cirrhosis Mortality. JAMA Netw. Open 2020, 3, e2023780. [Google Scholar] [CrossRef]
- Hart, A.; Wyatt, J. Evaluating Black-Boxes as Medical Decision Aids: Issues Arising from a Study of Neural Networks. Med. Inform. 1990, 15, 229–236. [Google Scholar] [CrossRef]
- Tu, J.V. Advantages and Disadvantages of Using Artificial Neural Networks versus Logistic Regression for Predicting Medical Outcomes. J. Clin. Epidemiol. 1996, 49, 1225–1231. [Google Scholar] [CrossRef]
- Paternostro, R.; Kapzan, L.; Mandorfer, M.; Schwarzer, R.; Benedikt, S.; Viveiros, A.; Bauer, D.; Ferlitsch, M.; Zoller, H.; Trauner, M.; et al. Anemia and Iron Deficiency in Compensated and Decompensated Cirrhosis: Prevalence and Impact on Clinical Outcomes. J. Gastroenterol. Hepatol. 2020, 35, 1619–1627. [Google Scholar] [CrossRef]
- Wan, S.-Z.; Nie, Y.; Zhang, Y.; Liu, C.; Zhu, X. Assessing the Prognostic Performance of the Child-Pugh, Model for End-Stage Liver Disease, and Albumin-Bilirubin Scores in Patients with Decompensated Cirrhosis: A Large Asian Cohort from Gastroenterology Department. Dis. Markers 2020, 2020, 5193028. [Google Scholar] [CrossRef]
- Godfrey, E.L.; Malik, T.H.; Lai, J.C.; Mindikoglu, A.L.; Galván, N.T.N.; Cotton, R.T.; O’Mahony, C.A.; Goss, J.A.; Rana, A. The Decreasing Predictive Power of MELD in an Era of Changing Etiology of Liver Disease. Am. J. Transplant. 2019, 19, 3299–3307. [Google Scholar] [CrossRef]


| Parameter | Characteristics | |
|---|---|---|
| n = 290 | Mean | Standard Deviation |
| Age (years) | 62.97 | 10.46 |
| WBC (×109/L) | 7.67 | 4.77 |
| Hemoglobin (g/L) | 92.34 | 30.63 |
| Platelets (×109/L) | 120 | 72 |
| Creatinine (μmol/L) | 125.10 | 92.75 |
| Total bilirubin (μmol/L) | 121.94 | 308.89 |
| INR | 1.61 | 0.49 |
| Sodium (mmol/L) | 135 | 6 |
| Albumin (g/L) | 26.81 | 5.87 |
| MELD-Na | 18 | 8 |
| MELD 3.0 | 21 | 8 |
| n | % | |
| Ascites | ||
| Grade 0 | 69 | 23.8 |
| Grade I | 186 | 64.1 |
| Grade II | 35 | 12.1 |
| Encephalopathy | ||
| Grade 0 | 191 | 65.9 |
| Grade I | 15 | 5.2 |
| Grade II | 47 | 16.2 |
| Grade III | 36 | 12.4 |
| Grade IV | 1 | 0.3 |
| Etiology of cirrhosis | ||
| Alcohol | 230 | 79.3 |
| HBV/HCV | 36 | 12.4 |
| Other + cryptogenic * | 12 | 4.1 |
| PBC/PSC | 7 | 2.4 |
| NAFLD | 5 | 1.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salkić, N.; Jovanović, P.; Barišić Jaman, M.; Selimović, N.; Paštrović, F.; Grgurević, I. Machine Learning for Short-Term Mortality in Acute Decompensation of Liver Cirrhosis: Better than MELD Score. Diagnostics 2024, 14, 981. https://doi.org/10.3390/diagnostics14100981
Salkić N, Jovanović P, Barišić Jaman M, Selimović N, Paštrović F, Grgurević I. Machine Learning for Short-Term Mortality in Acute Decompensation of Liver Cirrhosis: Better than MELD Score. Diagnostics. 2024; 14(10):981. https://doi.org/10.3390/diagnostics14100981
Chicago/Turabian StyleSalkić, Nermin, Predrag Jovanović, Mislav Barišić Jaman, Nedim Selimović, Frane Paštrović, and Ivica Grgurević. 2024. "Machine Learning for Short-Term Mortality in Acute Decompensation of Liver Cirrhosis: Better than MELD Score" Diagnostics 14, no. 10: 981. https://doi.org/10.3390/diagnostics14100981








