Clostridioides difficile and Enterococci’s Interplay in the Human Gut: Bacterial Alliance or Competition? A Systematic Literature Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy and Article Identification
2.2. Eligibility Criteria
2.3. Study Selection and Data Extraction
2.4. Data Synthesis
3. Results
3.1. Study Description
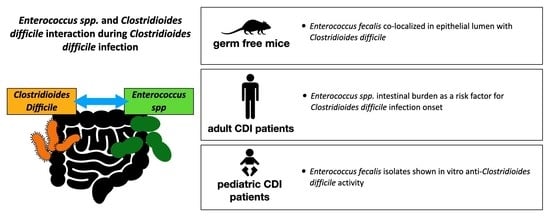
3.2. Enterococcus spp. and C. difficile’s Interplay during Clostridioides Difficile Infection
3.3. Enterococci Intestinal Burden as a Risk Factor for CDI
3.4. Vancomycin-Resistant Enterococci and Clostridioides difficile
3.4.1. Vancomycin-Resistant Enterococci Colonization as a Risk Factor for CDI
3.4.2. Effect of the CDI Antimicrobial Treatment on the Rate of Vancomycin-Resistant Enterococci Colonization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schubert, A.M.; Rogers, M.A.; Ring, C.; Mogle, J.; Petrosino, J.P.; Young, V.B.; Aronoff, D.M.; Schloss, P.D. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio 2014, 5, e01021-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auchtung, J.M.; Preisner, E.C.; Collins, J.; Lerma, A.I.; Britton, R.A. Identification of simplified microbial communities that inhibit Clostridioides difficile infection through dilution/extinction. mSphere 2020, 5, e00387-20. [Google Scholar] [CrossRef]
- Abbas, A.; Zackular, J.P. Microbe-microbe interactions during Clostridioides difficile infection. Curr. Opin. Microbiol. 2020, 53, 19–25. [Google Scholar] [CrossRef]
- Antharam, V.C.; Li, E.C.; Ishmael, A.; Sharma, A.; Mai, V.; Rand, K.H.; Wang, G.P. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 2013, 51, 2884–2892. [Google Scholar] [CrossRef] [Green Version]
- Poduval, R.D.; Kamath, R.P.; Corpuz, M.; Norkus, E.P.; Pitchumoni, C.S. Clostridium difficile and vancomycin-resistant Enterococcus: The new nosocomial alliance. Am. J. Gastroenterol. 2000, 95, 3513–3515. [Google Scholar] [CrossRef]
- Zacharioudakis, I.M.; Zervou, F.N.; Pliakos, E.E.; Ziakas, P.D.; Mylonakis, E. Colonization with toxinogenic C. difficile upon hospital admission, and risk of infection: A systematic review and meta-analysis. Am. J. Gastroenterol. 2015, 110, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Silva, V.; Dapkevicius, M.L.E.; Igrejas, G.; Poeta, P. Enterococci, from Harmless Bacteria to a Pathogen. Microorganisms 2020, 8, 1118. [Google Scholar] [CrossRef]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bäumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef] [Green Version]
- Bogdanová, K.; Doubravská, L.; Vágnerová, I.; Hricová, K.; Pudová, V.; Röderová, M.; Papajk, J.; Uvízl, R.; Langová, K.; Kolář, M. Clostridioides difficile and Vancomycin-Resistant Enterococci in COVID-19 Patients with Severe Pneumonia. Life 2021, 11, 1127. [Google Scholar] [CrossRef]
- Shin, J.W.; Yong, D.; Kim, M.S.; Chang, K.H.; Lee, K.; Kim, J.M.; Chong, Y. Sudden increase of vancomycin-resistant enterococcal infections in a Korean tertiary care hospital: Possible consequences of increased use of oral vancomycin. J. Infect. Chemother. 2003, 9, 62–67, discussion 104–105. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, N.A.; Schubert, A.M.; Flynn, K.J.; Leslie, J.L.; Sinani, H.; Bergin, I.L.; Young, V.B.; Schloss, P.D. The Gut Bacterial Community Potentiates Clostridioides difficile Infection Severity. mBio 2022, 13, e0118322. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.B.; Jenior, M.L.; Keenan, O.; Hart, J.L.; Specker, J.; Abbas, A.; Rangel, P.C.; Di, C.; Green, J.; Bustin, K.A.; et al. Enterococci enhance Clostridioides difficile pathogenesis. Nature 2022, 611, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Romyasamit, C.; Thatrimontrichai, A.; Aroonkesorn, A.; Chanket, W.; Ingviya, N.; Saengsuwan, P.; Singkhamanan, K. Enterococcus faecalis Isolated From Infant Feces Inhibits Toxigenic Clostridioides (Clostridium) difficile. Front. Pediatr. 2020, 8, 572633. [Google Scholar] [CrossRef]
- van Werkhoven, C.H.; Ducher, A.; Berkell, M.; Mysara, M.; Lammens, C.; Torre-Cisneros, J.; Rodríguez-Baño, J.; Herghea, D.; Cornely, O.A.; Biehl, L.M.; et al. Incidence and predictive biomarkers of Clostridioides difficile infection in hospitalized patients receiving broad-spectrum antibiotics. Nat. Commun. 2021, 12, 2240. [Google Scholar] [CrossRef]
- Berkell, M.; Mysara, M.; Xavier, B.B.; van Werkhoven, C.H.; Monsieurs, P.; Lammens, C.; Ducher, A.; Vehreschild, M.J.G.T.; Goossens, H.; de Gunzburg, J.; et al. Microbiota-based markers predictive of development of Clostridioides difficile infection. Nat. Commun. 2021, 12, 2241. [Google Scholar] [CrossRef]
- Vakili, B.; Fateh, A.; Asadzadeh Aghdaei, H.; Sotoodehnejadnematalahi, F.; Siadat, S.D. Characterization of Gut Microbiota in Hospitalized Patients with Clostridioides difficile Infection. Curr. Microbiol. 2020, 77, 1673–1680. [Google Scholar] [CrossRef]
- Axelrad, J.E.; Lebwohl, B.; Cuaresma, E.; Cadwell, K.; Green, P.H.R.; Freedberg, D.E. Gut colonization with vancomycin-resistant Enterococcus and risk for subsequent enteric infection. Gut Pathog. 2018, 10, 28. [Google Scholar] [CrossRef] [Green Version]
- Tickler, I.A.; Dela Cruz, C.M.; Obradovich, A.E.; Goering, R.V.; Dewell, S.; Le, V.M.; Tenover, F.C.; Healthcare Associated Infections Consortium. Presence of Clostridioides difficile and multidrug-resistant healthcare-associated pathogens in stool specimens from hospitalized patients in the USA. J. Hosp. Infect. 2020, 106, 179–185. [Google Scholar] [CrossRef]
- Kuzma, J.; Palcová, L.; Timko, J.; Bastová, V.; Janošcová, V.; Chmelař, D. Detection and molecular characterization of VRE isolates in Slovakia from stool samples positive for Clostridioides difficile toxins. Folia Microbiol. 2022, 67, 975–984. [Google Scholar] [CrossRef]
- Fishbein, S.R.S.; Hink, T.; Reske, K.A.; Cass, C.; Struttmann, E.; Iqbal, Z.H.; Seiler, S.; Kwon, J.H.; Burnham, C.A.; Dantas, G.; et al. Randomized Controlled Trial of Oral Vancomycin Treatment in Clostridioides difficile-Colonized Patients. mSphere 2021, 6, e00936-20. [Google Scholar] [CrossRef] [PubMed]
- Stevens, V.W.; Khader, K.; Echevarria, K.; Nelson, R.E.; Zhang, Y.; Jones, M.; Timbrook, T.T.; Samore, M.H.; Rubin, M.A. Use of Oral Vancomycin for Clostridioides difficile Infection and the Risk of Vancomycin-Resistant Enterococci. Clin. Infect. Dis. 2020, 71, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Correa-Martínez, C.L.; Hagemeier, N.C.J.; Froböse, N.J.; Kampmeier, S. Impact of Clostridioides difficile Therapy on Nosocomial Acquisition of Vancomycin-Resistant Enterococci. Pharmaceuticals 2021, 14, 1066. [Google Scholar] [CrossRef] [PubMed]
- Nerandzic, M.M.; Mullane, K.; Miller, M.A.; Babakhani, F.; Donskey, C.J. Reduced acquisition and overgrowth of vancomycin-resistant enterococci and Candida species in patients treated with fidaxomicin versus vancomycin for Clostridium difficile infection. Clin. Infect. Dis. 2012, 55, S121–S126. [Google Scholar] [CrossRef] [PubMed]

| Author, Year and Country | Study Type | Setting | Study Population, Age (Mean) and Sex (% Male) | Study Aim | Study Design | Study Results |
|---|---|---|---|---|---|---|
| Lesniak NA et al., 2022, US [12] | Retrospective cohort study in the animal model | University germ-free mouse core laboratory | Germfree C57BL/6 mice | To test the hypothesis that specific gut bacterial communities determine variation in CDI severity. | Various gut communities were derived by colonizing germfree mice with different human fecal communities. The mice were then infected with a single C. difficile ribotype 027 clinical isolate. | A variation in severity was observed according to each administered human fecal community. Communities rich in Enterococcus spp. determined more severe CDI. |
| Smith AB et al., 2022, US [13] | Retrospective cohort study in the animal model and in the human model | Three university hospitals from September 2015 through December 2019 | Animal model study: male mice. Retrospective cohort study: pediatric patients with median age (IQR) of 13 (5–16) years and with 50% male patients | To define the interaction between Enterococcus spp. and C. difficile. To quantify Enterococcus spp. burden in pediatric patients with CDI. | Mice were infected with toxigenic and nontoxigenic C. difficile strains following antibiotic-mediated depletion of endogenous Enterococcus spp. Fluorescent in situ hybridization was performed during CDI in mice. Stool samples were collected from pediatric CDI patients and were evaluated through metabolomic analyses. | E. faecalis burden was significantly increased in the presence of C. difficile toxin. Enterococcus faecalis colocalized with C. difficile in the lumen and in biofilm-like aggregates on the host epithelium. There was an enrichment of Enterococcus spp. in the stool of pediatric CDI patients, with positive correlation between Enterococcus spp. and C. difficile burdens (Spearman’s correlation: 0.551). |
| Romyasamit C et al., 2020, Thailand [14] | Microbiology study | One hospital | In total, 38 breast-fed healthy infants | To identify potential probiotic Enterococcus spp. strains exerting a protective effect towards CDI. | Agar well-diffusion assay was used to test the inhibitory activity of isolated colonies against toxigenic C. difficile strains. The cytopathic effects of C. difficile on colon adenocarcinoma cells were evaluated through immunofluorescence assay. | In total, 85 distinct bacterial colonies were isolated from the feces of 38 breast-fed infants. Of these, six Enterococcus faecalis isolates showed anti-C. difficile activity. The six strains inhibited spore germination (100 − 98.20 ± 2.17%) and sporulation. The cell-free supernatant of these strains reduced the cytopathic effects of C. difficile on colon adenocarcinoma cells (HT-29 cells). |
| Author, Year and Country | Study Type | Setting | Study Population, Age (Mean) and Sex (% Male) | Study Aim | Study Design | Study Results |
|---|---|---|---|---|---|---|
| van Werkhoven CH et al. ANTICIPATE Study Group, 2021, Europe [15] | International multicenter prospective observational cohort study | In total, 34 hospitals from France, Germany, Greece, the Netherlands, Romania and Spain from September 2016 through October 2017 | In total, 1007 patients receiving newly initiated antibiotic treatment with a median age of 70 years (IQR: 62–79) and 592 (58.8%) males | To assess CDI incidence among patients receiving antibiotic treatment. To assess clinical characteristic and biomarkers to predict CDI. | Patients receiving antibiotic treatment with penicillins, cephalosporins, carbapenems, fluoroquinolones or clindamycin were followed up for 90 days. If participants reported diarrhea during the follow-up, a fecal sample was collected and tested for CDI. | The estimated cumulative incidence of CDI was 1.1% (95% CI: 0.6–2.1) within 28 days and 1.9% (95% CI: 1.1–3.0) within 90 days. High intestinal abundance of Enterococcus spp. relative to Ruminococcus spp. (hazard ratio (95% CI): 5.4 (2.1–18.7)) and low Shannon alpha diversity index (9.7 (3.2–29.7)) predicted an increased CDI risk. |
| Berkell M et al. ANTICIPATE study group. 2021, Europe [16] | Multicenter observational prospective study | In total, 34 hospitals from France, Germany, Greece, the Netherlands, Romania and Spain from September 2016 through October 2017 | In total, 1007 patients receiving newly initiated antibiotic treatment with a median age of 70 years (IQR: 62–79) and 592 (58.8%) males. Of them, 15 were diagnosed with CDI | To identify microbial markers predictive of CDI and antibiotic-associated diarrhea. | Intestinal microbiota of the patients was investigated through 16S rRNA gene profiling combined with high-resolution sequence typing. | Patients developing CDI had significantly lower microbial diversity prior to antibiotic treatment and a microbiota enriched in Enterococcus spp. and depleted of Ruminococcus, Blautia, Prevotella and Bifidobacterium spp. Alpha diversity index was lower in patients developing CDI (p ≤ 0.049). |
| Vakili B et al., 2020, Iran [17] | Case-control study | Single hospital between 2019 and 2020 | In total, 50 inpatients with CDI and 50 healthy persons | To evaluate the composition of the gut microbiota in CDI patients compared to healthy control subjects. | C. difficile isolates were characterized through anaerobic culture and multiplex PCR. | Higher relative abundance of Enterococcus spp. in the CDI group compared to the healthy control group (p < 0.05). |
| Schubert AM et al., 2014, US [1] | Retrospective cohort study | One university hospital from October 2010 to January 2012 | In total, 338 individuals, including 94 CDI cases, 89 diarrheal controls and 155 nondiarrheal controls | To assess clinical and microbiome-based factors associated with CDI. | 16S rRNA gene sequencing was used to characterize the gut microbiomes. Clinical and microbiome data were merged to generate models of CDI status in order to differentiate between the three groups of subjects (CDI cases and diarrheal and nondiarrheal controls). | Subjects with CDI were significantly more likely to harbor Enterococcus spp. in comparison to nondiarrheal controls. |
| Author, Year and Country | Study Type | Setting | Study Population, Age (Mean) Sex (% Male) | Study Aim | Study Design | Study Results |
|---|---|---|---|---|---|---|
| Axelrad JE et al., 2018, US [18] | Multicenter retrospective cohort study | Nine intensive care units within two hospitals between 2012 and 2017 | In total, 716 patients. Of them, 131 were colonized with VRE, and 57 (43.5%) were male | To evaluate the risk, risk factors and pathogenic distribution of enteric infections in patients colonized with VRE. | Patients were screened for VRE colonization via rectal swab and culture. CDI was diagnosed through multiplex PCR assay. | A trend towards more CDIs was reported in patients colonized with VRE (15% vs. 10%, p: 0.11). |
| Tickler IA et al., 2020, US [19] | Microbiology study | Nine consortium laboratories between 2017 and 2018 | In total, 10 CDI patients and 9 non-CDI patients, providing a total of 363 stool samples | To investigate the presence of MDR bacteria in stools from CDI and non-CDI hospitalized patients. | Stool samples were analyzed through culture, MALDI-TOF mass spectrometry, PCR testing and whole-genome sequencing. | Among 363 stool samples, 175 yielded toxigenic C. difficile isolates. No significant differences were observed in VRE rates between CDI and non-CDI samples. |
| Kuzma J et al., 2022, Czech Republic [20] | Microbiology study | One military hospital from July 2020 to September 2021 | In total, 113 stool samples positive for C. difficile toxins | To identify VRE-colonized patients among hospitalized patients with CDI. | Stool samples were grown in a brain–heart infusion medium under aerobic conditions. The samples for VRE identification were grown on agar. The presence of vanA/vanB genes was tested through PCR. | Out of 113 samples, 44 (38.9%) harbored VRE. The most prevalent isolates were E. faecium (62%), E. faecalis (21%) and E. solitarius (9%). |
| Author, Year and Country | Study Type | Setting | Study Population, Age (Mean) Sex (% Male) | Study Aim | Study Design | Study Results |
|---|---|---|---|---|---|---|
| Fishbein SRS et al., 2021, US [21] | Double-blind randomized phase III controlled trial | One hospital from November 2017 to January 2019 | In total, 15 patients with at least one diarrheal stool testing positive for C. difficile through PCR but negative for C. difficile toxins via enzyme immunoassays with a median age of 66 (37–81) and 5 males (33.3%) | To examine the effect of oral vancomycin on the gut microbiota of hospitalized patients. | Patients were randomized 1:1 to receive 10 days of oral vancomycin (125 mg, 4 times per day) versus matching placebo. Stool collection and environmental sampling were performed at enrollment, day 5, day 10, week 4 and week 8 for bacterial culturing, metagenomic analysis and whole-genome sequencing analysis. | In the vancomycin-treated group, beta diversity (p: 0.0059) increased after the oral vancomycin treatment. No significant difference in the prevalence of VRE colonization was observed between the two study arms. |
| Stevens VW et al., 2020, US [22] | Retrospective cohort study | Department of Veterans Affairs Health between 2006 and 2016 | In total, 15,780 CDI patients; 5267 CDI patients treated with vancomycin alone were matched to one or more metronidazole-treated patient with a median age at CDI diagnosis of 69.0 and 94.7% male patients | To evaluate the risk of VRE following oral vancomycin or metronidazole treatment among CDI patients. | CDI patients were included if they were treated with metronidazole or oral vancomycin and had no history of VRE in the previous year. Patients were followed for VRE isolated from a clinical culture within 3 months. | Patients treated with oral vancomycin were no more likely to develop VRE within 3 months than metronidazole-treated patients (adjusted relative risk of 0.96; 95% CI: 0.77 to 1.20), and there was an absolute risk difference of −0.11% (95% CI: −0.68% to 0.47%). |
| Correa-Martínez CL et al., 2021, Germany [23] | Retrospective cohort study | One 1427-bed tertiary care center between 2018 and 2020 | In total, 170 hospitalized CDI patients with a median age of 53 years and 94 male patients (55%) | To compare VRE colonization rates between previously VRE-negative patients receiving either metronidazole or oral vancomycin as a CDI treatment. | VRE status of CDI patients receiving treatment with metronidazole or oral vancomycin was assessed at the beginning of CDI treatment. VRE isolates collected from CDI patients were subjected to whole-genome sequencing. | In total, 14 patients (3 metronidazole-treated patients; 11 vancomycin-treated patients) acquired VRE after the CDI antimicrobial treatment. There were no significant differences in VRE acquisition rates between patients that received metronidazole and those treated with oral vancomycin (p: 0.98). |
| Nerandzic MM et al., 2012, US and Canada [24] | Double-blind randomized phase III controlled trial | Multiple hospitals | Of 548 total patients enrolled in the trial, 301 (55%) had stool samples available both prior to and at completion of CDI therapy: 160 vancomycin-treated patients and 141 fidaxomicin-treated patients | To test the hypothesis that fidaxomicin promotes VRE colonization less than vancomycin. | CDI patients were randomized to receive treatment with 10 days of fidaxomicin versus 10 days of vancomycin. Pre- and posttreatment stool samples were collected and assessed for VRE colonization. | In comparison with vancomycin-treated patients, fidaxomicin-treated patients had less frequent acquisition of VRE (8 of 114 patients (7%) vs. 41 of 133 patients (31%); p < 0.001). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Granata, G.; Schiavone, F.; Taglietti, F.; Petrosillo, N. Clostridioides difficile and Enterococci’s Interplay in the Human Gut: Bacterial Alliance or Competition? A Systematic Literature Review. J. Clin. Med. 2023, 12, 4997. https://doi.org/10.3390/jcm12154997
Granata G, Schiavone F, Taglietti F, Petrosillo N. Clostridioides difficile and Enterococci’s Interplay in the Human Gut: Bacterial Alliance or Competition? A Systematic Literature Review. Journal of Clinical Medicine. 2023; 12(15):4997. https://doi.org/10.3390/jcm12154997
Chicago/Turabian StyleGranata, Guido, Francesco Schiavone, Fabrizio Taglietti, and Nicola Petrosillo. 2023. "Clostridioides difficile and Enterococci’s Interplay in the Human Gut: Bacterial Alliance or Competition? A Systematic Literature Review" Journal of Clinical Medicine 12, no. 15: 4997. https://doi.org/10.3390/jcm12154997
APA StyleGranata, G., Schiavone, F., Taglietti, F., & Petrosillo, N. (2023). Clostridioides difficile and Enterococci’s Interplay in the Human Gut: Bacterial Alliance or Competition? A Systematic Literature Review. Journal of Clinical Medicine, 12(15), 4997. https://doi.org/10.3390/jcm12154997