Creation and Validation of Patient-Derived Cancer Model Using Peritoneal and Pleural Effusion in Patients with Advanced Ovarian Cancer: An Early Experience
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Information
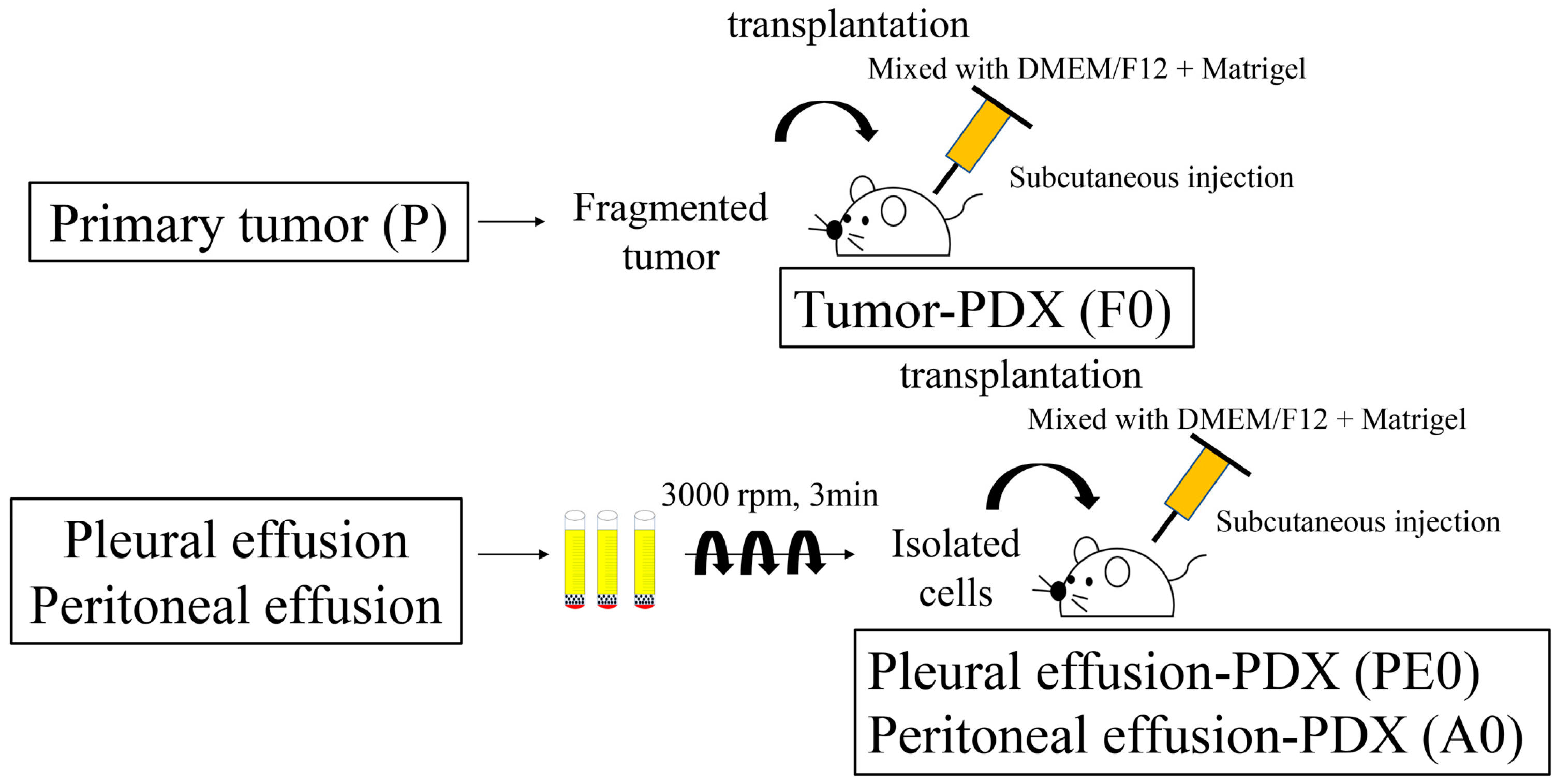
2.2. Tissue Samples
2.3. Establishment of Tumor-PDX (F0)
2.4. Establishment of PDX-BF (PE0/A0)
2.5. Animals
2.6. Sampling of the Xenograft Tumor
2.7. Pathological Analysis Using Immunohistochemistry
2.8. Sample Preparation for DNA and RNA Sequencing
2.9. Library Prepare
2.10. Run for DNA and RNA Sequencing
2.11. Data Analyses
2.12. Statistical Analyses
3. Results
3.1. Establishment of PDX Tumor
3.2. Histological Evaluation
3.3. Gene Mutation Analysis
3.4. Gene Expression Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, H.; Jiao, D.; Liu, A.; Wu, K. Tumor organoids: Applications in cancer modeling and potentials in precision medicine. J. Hematol. Oncol. 2022, 15, 58. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, M.; Wabel, E.; Mitchell, K.; Horibata, S. Mechanisms of chemotherapy resistance in ovarian cancer. Cancer Drug Resist. 2022, 5, 304–316. [Google Scholar] [CrossRef]
- Pan, K.; Gong, J.; Huynh, K.; Cristea, M. Current Systemic Treatment Landscape of Advanced Gynecologic Malignancies. Target. Oncol. 2019, 14, 269–283. [Google Scholar] [CrossRef]
- Nunes, M.; Ricardo, S. Chemoresistance in Ovarian Cancer: The Role of Malignant Ascites. In Ovarian Cancer; Lele, S., Ed.; Exon Publications: Brisbane, AU, Australia, 2022. [Google Scholar]
- Ayantunde, A.A.; Parsons, S.L. Pattern and prognostic factors in patients with malignant ascites: A retrospective study. Ann. Oncol. 2007, 18, 945–949. [Google Scholar] [CrossRef]
- Chen, J.; Wei, Z.; Fu, K.; Duan, Y.; Zhang, M.; Li, K.; Guo, T.; Yin, R. Non-apoptotic cell death in ovarian cancer: Treatment, resistance and prognosis. Biomed. Pharmacother. 2022, 150, 112929. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Dumitru, A.; Dobrica, E.C.; Croitoru, A.; Cretoiu, S.M.; Gaspar, B.S. Focus on PD-1/PD-L1 as a Therapeutic Target in Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 12067. [Google Scholar] [CrossRef]
- Kietpeerakool, C.; Rattanakanokchai, S.; Jampathong, N.; Srisomboon, J.; Lumbiganon, P. Management of drainage for malignant ascites in gynaecological cancer. Cochrane Database Syst. Rev. 2019, 12, CD007794. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, Z.H.; Wang, S.; Lang, J.H. Circulating Cell-Free DNA or Circulating Tumor DNA in the Management of Ovarian and Endometrial Cancer. Onco Targets Ther. 2019, 12, 11517–11530. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Nishie, R.; Ueda, S.; Miyamoto, S.; Hashida, S.; Konishi, H.; Terada, S.; Kogata, Y.; Sasaki, H.; Tsunetoh, S.; et al. Patient-Derived Xenograft Models in Cervical Cancer: A Systematic Review. Int. J. Mol. Sci. 2021, 22, 9369. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Nishie, R.; Ueda, S.; Miyamoto, S.; Hashida, S.; Konishi, H.; Terada, S.; Kogata, Y.; Sasaki, H.; Tsunetoh, S.; et al. Endometrial Cancer Patient-Derived Xenograft Models: A Systematic Review. J. Clin. Med. 2022, 11, 2606. [Google Scholar] [CrossRef] [PubMed]
- Kuwata, T.; Yanagihara, K.; Iino, Y.; Komatsu, T.; Ochiai, A.; Sekine, S.; Taniguchi, H.; Katai, H.; Kinoshita, T.; Ohtsu, A. Establishment of Novel Gastric Cancer Patient-Derived Xenografts and Cell Lines: Pathological Comparison between Primary Tumor, Patient-Derived, and Cell-Line Derived Xenografts. Cells 2019, 8, 585. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.M.; Jung, S.H.; Chung, Y.J. Comparison of the Genetic Alterations between Primary Colorectal Cancers and Their Corresponding Patient-Derived Xenograft Tissues. Genom. Inform. 2018, 16, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shen, C.; Wei, Z.; Zhang, R.; Wang, Y.; Jiang, L.; Chen, K.; Qiu, S.; Zhang, Y.; Zhang, T.; et al. Patient-derived non-small cell lung cancer xenograft mirrors complex tumor heterogeneity. Cancer Biol. Med. 2021, 18, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Abel, L.; Durmaz, A.; Hu, R.; Longhurst, C.; Baschnagel, A.M.; Wheeler, D.; Scott, J.G.; Kimple, R.J. Impact of immediate cryopreservation on the establishment of patient derived xenografts from head and neck cancer patients. J. Transl. Med. 2021, 19, 180. [Google Scholar] [CrossRef]
- Takayanagi, D.; Hirose, S.; Kuno, I.; Asami, Y.; Murakami, N.; Matsuda, M.; Shimada, Y.; Sunami, K.; Komatsu, M.; Hamamoto, R.; et al. Comparative Analysis of Genetic Alterations, HPV-Status, and PD-L1 Expression in Neuroendocrine Carcinomas of the Cervix. Cancers 2021, 13, 1215. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Tanaka, T.; Hirosuna, K.; Nishie, R.; Ueda, S.; Hashida, S.; Terada, S.; Konishi, H.; Kogata, Y.; Taniguchi, K.; et al. Validation of a Patient-Derived Xenograft Model for Cervical Cancer Based on Genomic and Phenotypic Characterization. Cancers 2022, 14, 2969. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Xue, H.; Sutcliffe, M.; Gout, P.W.; Huntsman, D.G.; Miller, D.M.; Gilks, C.B.; Wang, Y.Z. Establishment of subrenal capsule xenografts of primary human ovarian tumors in SCID mice: Potential models. Gynecol. Oncol. 2005, 96, 48–55. [Google Scholar] [CrossRef]
- Dong, R.; Qiang, W.; Guo, H.; Xu, X.; Kim, J.J.; Mazar, A.; Kong, B.; Wei, J.J. Histologic and molecular analysis of patient derived xenografts of high-grade serous ovarian carcinoma. J. Hematol. Oncol. 2016, 9, 92. [Google Scholar] [CrossRef]
- Heo, E.J.; Cho, Y.J.; Cho, W.C.; Hong, J.E.; Jeon, H.K.; Oh, D.Y.; Choi, Y.L.; Song, S.Y.; Choi, J.J.; Bae, D.S.; et al. Patient-Derived Xenograft Models of Epithelial Ovarian Cancer for Preclinical Studies. Cancer Res. Treat. 2017, 49, 915–926. [Google Scholar] [CrossRef]
- Cybula, M.; Wang, L.; Wang, L.; Drumond-Bock, A.L.; Moxley, K.M.; Benbrook, D.M.; Gunderson-Jackson, C.; Ruiz-Echevarria, M.J.; Bhattacharya, R.; Mukherjee, P.; et al. Patient-Derived Xenografts of High-Grade Serous Ovarian Cancer Subtype as a Powerful Tool in Pre-Clinical Research. Cancers 2021, 13, 6288. [Google Scholar] [CrossRef]
- Dobbin, Z.C.; Katre, A.A.; Steg, A.D.; Erickson, B.K.; Shah, M.M.; Alvarez, R.D.; Conner, M.G.; Schneider, D.; Chen, D.; Landen, C.N. Using heterogeneity of the patient-derived xenograft model to identify the chemoresistant population in ovarian cancer. Oncotarget 2014, 5, 8750–8764. [Google Scholar] [CrossRef]
- Ricci, F.; Bizzaro, F.; Cesca, M.; Guffanti, F.; Ganzinelli, M.; Decio, A.; Ghilardi, C.; Perego, P.; Fruscio, R.; Buda, A.; et al. Patient-derived ovarian tumor xenografts recapitulate human clinicopathology and genetic alterations. Cancer Res. 2014, 74, 6980–6990. [Google Scholar] [CrossRef]
- Topp, M.D.; Hartley, L.; Cook, M.; Heong, V.; Boehm, E.; McShane, L.; Pyman, J.; McNally, O.; Ananda, S.; Harrell, M.; et al. Molecular correlates of platinum response in human high-grade serous ovarian cancer patient-derived xenografts. Mol. Oncol. 2014, 8, 656–668. [Google Scholar] [CrossRef]
- Li, L.Y.; Kim, H.J.; Park, S.A.; Lee, S.H.; Kim, L.K.; Lee, J.Y.; Kim, S.; Kim, Y.T.; Kim, S.W.; Nam, E.J. Genetic Profiles Associated with Chemoresistance in Patient-Derived Xenograft Models of Ovarian Cancer. Cancer Res. Treat. 2019, 51, 1117–1127. [Google Scholar] [CrossRef]
- Ben-David, U.; Ha, G.; Tseng, Y.Y.; Greenwald, N.F.; Oh, C.; Shih, J.; McFarland, J.M.; Wong, B.; Boehm, J.S.; Beroukhim, R.; et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet. 2017, 49, 1567–1575. [Google Scholar] [CrossRef]
- Shi, J.; Li, Y.; Jia, R.; Fan, X. The fidelity of cancer cells in PDX models: Characteristics, mechanism and clinical significance. Int. J. Cancer 2019, 146, 2078–2088. [Google Scholar] [CrossRef]
- Golan, T.; Stossel, C.; Schvimer, M.; Atias, D.; Halperin, S.; Buzhor, E.; Raitses-Gurevich, M.; Cohen, K.; Pri-Chen, S.; Wilson, J.; et al. Pancreatic cancer ascites xenograft–an expeditious model mirroring advanced therapeutic resistant disease. Oncotarget 2017, 8, 40778–40790. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Lee, J.Y.; Lee, S.; Kim, D.; Lim, J.; Jun, H.R.; Jeon, S.; Kim, Y.A.; Park, H.S.; Kim, K.P.; et al. Establishing Patient-Derived Cancer Cell Cultures and Xenografts in Biliary Tract Cancer. Cancer Res. Treat. 2022, 55, 219–230. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, F.; Pan, X.; Wang, G.; Zhu, L.; Zhang, J.; Wen, D.; Lu, S. Xenograft tumors derived from malignant pleural effusion of the patients with non-small-cell lung cancer as models to explore drug resistance. Cancer Commun. 2018, 38, 19. [Google Scholar] [CrossRef]
- Kreuzinger, C.; Gamperl, M.; Wolf, A.; Heinze, G.; Geroldinger, A.; Lambrechts, D.; Boeckx, B.; Smeets, D.; Horvat, R.; Aust, S.; et al. Molecular characterization of 7 new established cell lines from high grade serous ovarian cancer. Cancer Lett. 2015, 362, 218–228. [Google Scholar] [CrossRef]
- Li, X.; Zhu, D.; Li, N.; Yang, H.; Zhao, Z.; Li, M. Characterization of ascites-derived tumor cells from an endometrial cancer patient. Cancer Sci. 2017, 108, 2352–2357. [Google Scholar] [CrossRef]
- Liu, J.F.; Palakurthi, S.; Zeng, Q.; Zhou, S.; Ivanova, E.; Huang, W.; Zervantonakis, I.K.; Selfors, L.M.; Shen, Y.; Pritchard, C.C.; et al. Establishment of Patient-Derived Tumor Xenograft Models of Epithelial Ovarian Cancer for Preclinical Evaluation of Novel Therapeutics. Clin. Cancer Res. 2017, 23, 1263–1273. [Google Scholar] [CrossRef]
- Verschraegen, C.F.; Hu, W.; Du, Y.; Mendoza, J.; Early, J.; Deavers, M.; Freedman, R.S.; Bast, R.C., Jr.; Kudelka, A.P.; Kavanagh, J.J.; et al. Establishment and characterization of cancer cell cultures and xenografts derived from primary or metastatic Mullerian cancers. Clin. Cancer Res. 2003, 9, 845–852. [Google Scholar]
- Press, J.Z.; Kenyon, J.A.; Xue, H.; Miller, M.A.; De Luca, A.; Miller, D.M.; Huntsman, D.G.; Gilks, C.B.; McAlpine, J.N.; Wang, Y.Z. Xenografts of primary human gynecological tumors grown under the renal capsule of NOD/SCID mice show genetic stability during serial transplantation and respond to cytotoxic chemotherapy. Gynecol. Oncol. 2008, 110, 256–264. [Google Scholar] [CrossRef]
- Cybula, M.; Bieniasz, M. Patient-derived tumor models are attractive tools to repurpose drugs for ovarian cancer treatment: Pre-clinical updates. Oncotarget 2022, 13, 553–575. [Google Scholar] [CrossRef]
- Machinaga, A.; Hori, Y.; Shimizu, K.; Okahara, K.; Yanagita, E.; Miyoshi, M.; Itoh, T.; Sasai, K. Xenografts Derived From Patients’ Ascites Recapitulate the Gemcitabine Resistance Observed in Pancreatic Cancer Patients. Pancreas 2019, 48, 1294–1302. [Google Scholar] [CrossRef]
- Lee, A.Q.; Ijiri, M.; Rodriguez, R.; Gandour-Edwards, R.; Lee, J.; Tepper, C.G.; Li, Y.; Beckett, L.; Lam, K.; Goodwin, N.; et al. Novel Patient Metastatic Pleural Effusion-Derived Xenograft Model of Renal Medullary Carcinoma Demonstrates Therapeutic Efficacy of Sunitinib. Front. Oncol. 2021, 11, 648097. [Google Scholar] [CrossRef]
- Weroha, S.J.; Becker, M.A.; Enderica-Gonzalez, S.; Harrington, S.C.; Oberg, A.L.; Maurer, M.J.; Perkins, S.E.; AlHilli, M.; Butler, K.A.; McKinstry, S.; et al. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clin. Cancer Res. 2014, 20, 1288–1297. [Google Scholar] [CrossRef]
- Cavalloni, G.; Peraldo-Neia, C.; Sassi, F.; Chiorino, G.; Sarotto, I.; Aglietta, M.; Leone, F. Establishment of a patient-derived intrahepatic cholangiocarcinoma xenograft model with KRAS mutation. BMC Cancer 2016, 16, 90. [Google Scholar] [CrossRef] [PubMed]
- Ojima, H.; Yamagishi, S.; Shimada, K.; Shibata, T. Establishment of various biliary tract carcinoma cell lines and xenograft models for appropriate preclinical studies. World J. Gastroenterol. 2016, 22, 9035–9038. [Google Scholar] [CrossRef]
- Glaser, G.; Weroha, S.J.; Becker, M.A.; Hou, X.; Enderica-Gonzalez, S.; Harrington, S.C.; Haluska, P. Conventional chemotherapy and oncogenic pathway targeting in ovarian carcinosarcoma using a patient-derived tumorgraft. PLoS ONE 2015, 10, e0126867. [Google Scholar] [CrossRef]
- Sakamoto, H.; Yamasaki, T.; Sumiyoshi, T.; Takeda, M.; Shibasaki, N.; Utsunomiya, N.; Arakaki, R.; Akamatsu, S.; Kobayashi, T.; Inoue, T.; et al. Functional and genomic characterization of patient-derived xenograft model to study the adaptation to mTORC1 inhibitor in clear cell renal cell carcinoma. Cancer Med. 2021, 10, 119–134. [Google Scholar] [CrossRef]
- Chaudary, N.; Pintilie, M.; Schwock, J.; Dhani, N.; Clarke, B.; Milosevic, M.; Fyles, A.; Hill, R.P. Characterization of the Tumor-Microenvironment in Patient-Derived Cervix Xenografts (OCICx). Cancers 2012, 4, 821–845. [Google Scholar] [CrossRef]
- Serebrenik, A.A.; Argyris, P.P.; Jarvis, M.C.; Brown, W.L.; Bazzaro, M.; Vogel, R.I.; Erickson, B.K.; Lee, S.H.; Goergen, K.M.; Maurer, M.J.; et al. The DNA Cytosine Deaminase APOBEC3B is a Molecular Determinant of Platinum Responsiveness in Clear Cell Ovarian Cancer. Clin. Cancer Res. 2020, 26, 3397–3407. [Google Scholar] [CrossRef]
- Cybulska, P.; Stewart, J.M.; Sayad, A.; Virtanen, C.; Shaw, P.A.; Clarke, B.; Stickle, N.; Bernardini, M.Q.; Neel, B.G. A Genomically Characterized Collection of High-Grade Serous Ovarian Cancer Xenografts for Preclinical Testing. Am. J. Pathol. 2018, 188, 1120–1131. [Google Scholar] [CrossRef]
- De Thaye, E.; Van de Vijver, K.; Van der Meulen, J.; Taminau, J.; Wagemans, G.; Denys, H.; Van Dorpe, J.; Berx, G.; Ceelen, W.; Van Bocxlaer, J.; et al. Establishment and characterization of a cell line and patient-derived xenograft (PDX) from peritoneal metastasis of low-grade serous ovarian carcinoma. Sci. Rep. 2020, 10, 6688. [Google Scholar] [CrossRef]
- Ricci, F.; Guffanti, F.; Affatato, R.; Brunelli, L.; Roberta, P.; Fruscio, R.; Perego, P.; Bani, M.R.; Chiorino, G.; Rinaldi, A.; et al. Establishment of patient-derived tumor xenograft models of mucinous ovarian cancer. Am. J. Cancer Res. 2020, 10, 572–580. [Google Scholar]
- Odunsi, A.; McGray, A.J.R.; Miliotto, A.; Zhang, Y.; Wang, J.; Abiola, A.; Eppolito, C.; Huang, R.Y. Fidelity of human ovarian cancer patient-derived xenografts in a partially humanized mouse model for preclinical testing of immunotherapies. J. Immunother. Cancer 2020, 8, e001237. [Google Scholar] [CrossRef]
- Choi, C.H.; Ryu, J.Y.; Cho, Y.J.; Jeon, H.K.; Choi, J.J.; Ylaya, K.; Lee, Y.Y.; Kim, T.J.; Chung, J.Y.; Hewitt, S.M.; et al. The anti-cancer effects of itraconazole in epithelial ovarian cancer. Sci. Rep. 2017, 7, 6552. [Google Scholar] [CrossRef] [PubMed]
- Serra, V.; Wang, A.T.; Castroviejo-Bermejo, M.; Polanska, U.M.; Palafox, M.; Herencia-Ropero, A.; Jones, G.N.; Lai, Z.; Armenia, J.; Michopoulos, F.; et al. Identification of a Molecularly-Defined Subset of Breast and Ovarian Cancer Models that Respond to WEE1 or ATR Inhibition, Overcoming PARP Inhibitor Resistance. Clin. Cancer Res. 2022, 28, 4536–4550. [Google Scholar] [CrossRef]
- West, H.J.; McCleland, M.; Cappuzzo, F.; Reck, M.; Mok, T.S.; Jotte, R.M.; Nishio, M.; Kim, E.; Morris, S.; Zou, W.; et al. Clinical efficacy of atezolizumab plus bevacizumab and chemotherapy in KRAS-mutated non-small cell lung cancer with STK11, KEAP1, or TP53 comutations: Subgroup results from the phase III IMpower150 trial. J. Immunother. Cancer. 2022, 10, e003027. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, Y.; Qian, L.; Wang, P. Emerging strategies to target RAS signaling in human cancer therapy. J. Hematol. Oncol. 2021, 14, 116. [Google Scholar] [CrossRef]
- Ji, W.; Niu, X.; Yu, Y.; Li, Z.; Gu, L.; Lu, S. SMO mutation predicts the effect of immune checkpoint inhibitor: From NSCLC to multiple cancers. Front. Immunol. 2022, 13, 955800. [Google Scholar] [CrossRef]
- Sun, Y.; Lu, D.; Yin, Y.; Song, J.; Liu, Y.; Hao, W.; Qi, F.; Zhang, G.; Zhang, X.; Liu, L.; et al. PTENalpha functions as an immune suppressor and promotes immune resistance in PTEN-mutant cancer. Nat. Commun. 2021, 12, 5147. [Google Scholar] [CrossRef]
- Asante, D.B.; Calapre, L.; Ziman, M.; Meniawy, T.M.; Gray, E.S. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: Ready for prime time? Cancer Lett. 2020, 468, 59–71. [Google Scholar] [CrossRef]
- Schwartz, M.; Camacho-Vanegas, O.; Wood, A.M.; Dashkoff, M.; Whitelock, C.; Harkins, T.T.; Cohen, C.J.; Beddoe, A.M.; Dottino, P.; Martignetti, J.A. Applying Precision Medicine to Ovarian Cancer: Proof-of-Principle for a “Molecular Second Look”. Int. J. Gynecol Cancer 2018, 28, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Barquin, M.; Maximiano, C.; Perez-Barrios, C.; Sanchez-Herrero, E.; Soriano, M.; Colmena, M.; Garcia-Espantaleon, M.; Tejerina Gonzalez, E.; Gutierrez, L.; Sanchez Ruiz, A.C.; et al. Peritoneal washing is an adequate source for somatic BRCA1/2 mutation testing in ovarian malignancies. Pathol. Res. Pract. 2019, 215, 392–394. [Google Scholar] [CrossRef] [PubMed]



| Overall | Established | Failed | p Value | ||
|---|---|---|---|---|---|
| Number of patients | 15 | 8 (53%) | 7 (47%) | ||
| Age *, years old | 64 (61–72) | 64 (62–66) | 71 (57–78) | 0.3 | |
| Histological type | high-grade serous carcinoma | 10 | 5 | 5 | |
| carcinosarcoma | 1 | 1 | 0 | ||
| low-grade serous carcinoma | 1 | 1 | 0 | ||
| clear cell carcinoma | 3 | 1 | 2 | ||
| FIGO stage | III | 7 | 3 | 4 | |
| IV | 8 | 5 | 3 | ||
| HRD positive | 6 | 3 | 3 | 1.0 | |
| BRCA positive | 1 | 0 | 1 | ||
| CA125 *, U/mL | 1138 (224–3117) | 865 (288–3344) | 1709 (115–3117) | 0.9 | |
| Follow-up *, months | 11 (10–18) | 11 (8–12) | 16 (11–20) | 0.03 | |
| PFS *, months | 10 (7–12) | 10 (7–11) | 11 (7–18) | 0.4 | |
| PDX Growth | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Case | Age | Type of Cancer | Histological Type | FIGO | HRD | Source | Tumor (F0) | Ascites (A0) | Pleural Effusion (PE0) |
| 1 | 65 | PC | high-grade serous carcinoma | 3C | positive (GIS 42) | ascites | Yes | Yes | - |
| 2 | 63 | PC | high-grade serous carcinoma | 4B | negative | ascites | Yes | Yes | - |
| 3 | 64 | OC | high-grade serous carcinoma | 4B | negative | ascites | Yes | Yes | - |
| 4 | 66 | PC | high-grade serous carcinoma | 4A | negative | ascites | Yes | Yes | - |
| 5 | 61 | OC | carcinosarcoma | 3C | positive (GIS 43) | ascites | No | Yes | - |
| 6 | 72 | OC | low-grade serous carcinoma | 3B | negative | ascites | No | Yes | - |
| 7 | 63 | OC | clear cell carcinoma | 3B | unknown | pleural effusion | Yes | - | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishie, R.; Tanaka, T.; Hirosuna, K.; Miyamoto, S.; Murakami, H.; Tsuchihashi, H.; Toji, A.; Ueda, S.; Morita, N.; Hashida, S.; et al. Creation and Validation of Patient-Derived Cancer Model Using Peritoneal and Pleural Effusion in Patients with Advanced Ovarian Cancer: An Early Experience. J. Clin. Med. 2024, 13, 2718. https://doi.org/10.3390/jcm13092718
Nishie R, Tanaka T, Hirosuna K, Miyamoto S, Murakami H, Tsuchihashi H, Toji A, Ueda S, Morita N, Hashida S, et al. Creation and Validation of Patient-Derived Cancer Model Using Peritoneal and Pleural Effusion in Patients with Advanced Ovarian Cancer: An Early Experience. Journal of Clinical Medicine. 2024; 13(9):2718. https://doi.org/10.3390/jcm13092718
Chicago/Turabian StyleNishie, Ruri, Tomohito Tanaka, Kensuke Hirosuna, Shunsuke Miyamoto, Hikaru Murakami, Hiromitsu Tsuchihashi, Akihiko Toji, Shoko Ueda, Natsuko Morita, Sousuke Hashida, and et al. 2024. "Creation and Validation of Patient-Derived Cancer Model Using Peritoneal and Pleural Effusion in Patients with Advanced Ovarian Cancer: An Early Experience" Journal of Clinical Medicine 13, no. 9: 2718. https://doi.org/10.3390/jcm13092718





