Screening for Antibiotic Resistance Genes in Bacteria and the Presence of Heavy Metals in the Upstream and Downstream Areas of the Wadi Hanifah Valley in Riyadh, Saudi Arabia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Extraction of Genomic DNA
2.3. Detection of Antibiotic-Resistant Genes (ARGs)
2.4. Determination of Heavy Metals
2.5. Quality Control
2.6. Statistical Analysis
3. Results
3.1. Heavy Metal Contents in Water Samples
3.2. Correlation between ARGs and Heavy Metals
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mendelson, M.; Matsoso, M.P. The World Health Organization global action plan for antimicrobial resistance. SAMJ S. Afr. Med. J. 2015, 105, 325. [Google Scholar] [CrossRef]
- Bank, W. Drug-Resistant Infections: A Threat to Our Economic Future; World Bank: Washington, DC, USA, 2017. [Google Scholar]
- Kim, D.W.; Cha, C.J. Antibiotic resistome from the One-Health perspective: Understanding and controlling antimicrobial resistance transmission. Exp. Mol. Med. 2021, 53, 301–309. [Google Scholar] [CrossRef]
- Ojo, O.E.; Fabusoro, E.; Majasan, A.A.; Dipeolu, M.A. Antimicrobials in animal production: Usage and practices among livestock farmers in Oyo and Kaduna States of Nigeria. Trop. Anim. Health Prod. 2016, 48, 189–197. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J. Antibiotic resistance genes from livestock waste: Occurrence, dissemination, and treatment. NPJ Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Serwecińska, L. Antimicrobials and antibiotic-resistant bacteria: A risk to the environment and to public health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Kaiser, R.A.; Taing, L.; Bhatia, H. Antimicrobial resistance and environmental health: A water stewardship framework for global and national action. Antibiotics 2022, 11, 63. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Petrovich, M.L.; Zilberman, A.; Kaplan, A.; Eliraz, G.R.; Wang, Y.; Langenfeld, K.; Wells, G.F. Microbial and viral communities and their antibiotic resistance genes throughout a hospital wastewater treatment system. Front. Microbiol. 2020, 11, 153. [Google Scholar] [CrossRef]
- Lu, Q.; Zhao, R.; Li, Q.; Ma, Y.; Chen, J.; Yu, Q.; An, S. Elemental composition and microbial community differences between wastewater treatment plant effluent and local natural surface water: A Zhengzhou city study. J. Environ. Manag. 2023, 325, 116398. [Google Scholar] [CrossRef]
- Grenni, P. Antimicrobial resistance in rivers: A review of the genes detected and new challenges. Environ. Toxicol. Chem. 2022, 41, 687–714. [Google Scholar] [CrossRef]
- Grenni, P.; Ancona, V.; Caracciolo, A.B. Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sànchez-Melsió, A.; Balcázar, J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef]
- Kümmerer, K. Significance of antibiotics in the environment. J. Antimicrob. Chemother. 2003, 52, 5–7. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- Sanseverino, I.; Navarro Cuenca, A.; Loos, R.; Marinov, D.; Lettieri, T. State of the Art on the Contribution of Water to Antimicrobial Resistance; Publications Office of the European Union: Luxembourg, 2018. [Google Scholar]
- Stange, C.; Yin, D.; Xu, T.; Guo, X.; Schäfer, C.; Tiehm, A. Distribution of clinically relevant antibiotic resistance genes in Lake Tai, China. Sci. Total Environ. 2019, 655, 337–346. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Berg, J.; Thorsen, M.K.; Holm, P.E.; Jensen, J.; Nybroe, O.; Brandt, K.K. Cu exposure under field conditions co-selects for antibiotic resistance as determined by a novel cultivation-independent bacterial community tolerance assay. Environ. Sci. Technol. 2010, 44, 8724–8728. [Google Scholar] [CrossRef]
- Chapman, J.S. Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int. Biodeterior. Biodegrad. 2003, 51, 271–276. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L.; Carrër, A.; Toleman, M.A.; Walsh, T.R. How to detect NDM-1 producers. J. Clin. Microbiol. 2011, 49, 718–721. [Google Scholar] [CrossRef]
- Pournajaf, A.; Ardebili, A.; Goudarzi, L.; Khodabandeh, M.; Narimani, T.; Abbaszadeh, H. PCR-based identification of methicillin–resistant Staphylococcus aureus strains and their antibiotic resistance profiles. Asian Pac. J. Trop. Biomed. 2014, 4, S293–S297. [Google Scholar] [CrossRef]
- Schwartz, T.; Kohnen, W.; Jansen, B.; Obst, U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 2023, 43, 325–335. [Google Scholar] [CrossRef]
- Aminov, R.I.; Garrigues-Jeanjean, N.; Mackie, R.I. Molecular Ecology of Tetracycline Resistance: Development and Validation of Primers for Detection of Tetracycline Resistance Genes Encoding Ribosomal Protection Proteins. Appl. Environ. Microbiol. 2001, 67, 22–32. [Google Scholar] [CrossRef]
- Gevers, D.; Danielsen, M.; Huys, G.; Swings, J. Molecular Characterization of tet (M) Genes in Lactobacillus Isolates from Different Types of Fermented Dry Sausage. Appl. Environ. Microbiol. 2003, 69, 1270–1275. [Google Scholar] [CrossRef]
- Vakulenko, S.B.; Donabedian, S.M.; Voskresenskiy, A.M.; Zervos, M.J.; Lerner, S.A.; Chow, J.W. Multiplex PCR for Detection of Aminoglycoside Resistance Genes in Enterococci. Antimicrob. Agents Chemother. 2003, 47, 1423–1426. [Google Scholar] [CrossRef]
- Maynard, C.; Fairbrother, J.M.; Bekal, S.; Sanschagrin, F.; Levesque, R.C.; Brousseau, R.; Masson, L.; Larivière, S.; Harel, J. Antimicrobial Resistance Genes in Enterotoxigenic Escherichia coli O149:K91 Isolates Obtained over a 23-Year Period from Pigs. Antimicrob. Agents Chemother. 2003, 47, 3214–3221. [Google Scholar] [CrossRef]
- Vassort-Bruneau, C.; Lesage-Descauses, M.-C.; Martel, J.-L.; Lafont, J.-P.; Chaslus-Dancla, E. CAT III chloramphenicol resistance in Pasteurella haemolytica and Pasteurella multocida isolated from calves. J. Antimicrob. Chemother. 1996, 38, 205–213. [Google Scholar] [CrossRef]
- Mishra, S.; Singh, A.K.; Cheng, L.; Hussain, A.; Maiti, A. Occurrence of antibiotics in wastewater: Potential ecological risk and removal through anaerobic–aerobic systems. Environ. Res. 2023, 226, 115678. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Komijani, M.; Shamabadi, N.S.; Shahin, K.; Eghbalpour, F.; Tahsili, M.R.; Bahram, M. Heavy metal pollution promotes antibiotic resistance potential in the aquatic environment. Environ. Pollut. 2021, 274, 116569. [Google Scholar] [CrossRef]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 2012, 3, 399. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, A.Z.; Cen, T.; Li, X.; He, M.; Li, D.; Chen, J. Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environ. Pollut. 2018, 237, 74–82. [Google Scholar] [CrossRef]
- Franklin, A.M.; Brinkman, N.E.; Jahne, M.A.; Keely, S.P. Twenty-first century molecular methods for analyzing antimicrobial resistance in surface waters to support One Health assessments. J. Microbiol. Methods 2021, 184, 106174. [Google Scholar] [CrossRef]
- Reichert, G.; Hilgert, S.; Alexander, J.; de Azevedo, J.C.R.; Morck, T.; Fuchs, S.; Schwartz, T. Determination of antibiotic resistance genes in a WWTP-impacted river in surface water, sediment, and biofilm: Influence of seasonality and water quality. Sci. Total Environ. 2021, 768, 144526. [Google Scholar] [CrossRef]
- Manoharan, R.K.; Ishaque, F.; Ahn, Y.H. Fate of antibiotic resistant genes in wastewater environments and treatment strategies—A review. Chemosphere 2022, 298, 134671. [Google Scholar] [CrossRef]
- Proia, L.; von Schiller, D.; Sànchez-Melsió, A.; Sabater, S.; Borrego, C.M.; Rodríguez-Mozaz, S.; Balcázar, J.L. Occurrence and persistence of antibiotic resistance genes in river biofilms after wastewater inputs in small rivers. Environ. Pollut. 2016, 210, 121–128. [Google Scholar] [CrossRef]
- Obayiuwana, A.; Ibekwe, A.M. Antibiotic resistance genes occurrence in wastewaters from selected pharmaceutical facilities in Nigeria. Water 2020, 12, 1897. [Google Scholar] [CrossRef]
- Kulik, K.; Lenart-Boroń, A.; Wyrzykowska, K. Impact of Antibiotic Pollution on the Bacterial Population within Surface Water with Special Focus on Mountain Rivers. Water 2023, 15, 975. [Google Scholar] [CrossRef]
- Yang, Y.; Niehaus, K.E.; Walker, T.M.; Iqbal, Z.; Walker, A.S.; Wilson, D.J.; Peto, T.E.A.; Crook, D.W.; Smith, E.G.; Zhu, T.; et al. Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data. Bioinformatics 2018, 34, 1666–1671. [Google Scholar] [CrossRef]
- El-Sayed Ahmed, M.A.E.G.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef]
- Silva, V.; Ferreira, E.; Manageiro, V.; Reis, L.; Tejedor-Junco, M.T.; Sampaio, A.; Poeta, P. Distribution and Clonal Diversity of Staphylococcus aureus and Other Staphylococci in Surface Waters: Detection of ST425-t742 and ST130-t843 mec C-Positive MRSA Strains. Antibiotics 2021, 10, 1416. [Google Scholar] [CrossRef]
- Andrade, F.F.; Silva, D.; Rodrigues, A.; Pina-Vaz, C. Colistin update on its mechanism of action and resistance, present and future challenges. Microorganisms 2020, 8, 1716. [Google Scholar] [CrossRef]
- World Health Organization. Guidelines for Drinking Water Quality, 4th ed.; incorporating the first addendum; World Health Organization: Geneva, Switzerland, 2017; pp. 1–631. [Google Scholar]
- World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- United States Environmental Protection Agency. Drinking Water Requirements for States and Public Water Systems, Chemical Contaminant Rules; United States Environmental Protection Agency: Washington, DC, USA, 2009. [Google Scholar]
- United States Environmental Protection Agency. Electronic Code of Federal Regulation (eCFR); Title 40: Protection of Environmen, Part 141—National Primary Drinking Water Regulations, Sub-part I: Control of Lead and Copper; United States Environmental Protection Agency: Washington, DC, USA, 2019. [Google Scholar]
- Wang, Y.; Li, B.L.; Zhu, J.L.; Feng, Q.; Liu, W.; He, Y.H.; Wang, X. Assessment of heavy metals in surface water, sediment and macrozoobenthos in inland rivers: A case study of the Heihe River, Northwest China. Environ. Sci. Pollut. Res. 2022, 29, 35253–35268. [Google Scholar] [CrossRef]
- Ji, X.; Shen, Q.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.; Wu, M. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China. J. Hazard. Mater. 2012, 235, 178–185. [Google Scholar] [CrossRef]
- Rajasekar, A.; Qiu, M.; Wang, B.; Murava, R.T.; Norgbey, E. Relationship between water quality, heavy metals and antibiotic resistance genes among three freshwater lakes. Environ. Monit. Assess. 2022, 194, 64. [Google Scholar] [CrossRef]



| Gene | Oligo Name | Oligo Sequence (5′ to 3′) | References |
|---|---|---|---|
| blaNDM-1 | NDM1-F | GGTTTGGCGATCTGGTTTTC | [22] |
| NDM1-R | CGGAATGGCTCATCACGATC | ||
| mecA | mecA F 1282 | AAAATCGATGGTAAAGGTTGGC | [23] |
| mecA R 1793 | AGTTCTGCAGTACCGGATTTGC | ||
| ampC | ampC-F | TTCTATCAAMACTGGCARCC | [24] |
| ampC-R | CCYTTTTATGTACCCAYGA | ||
| tet(M) | tet(M)-F | ACAGAAAGCTTATTATATAAC | [25] |
| tet(M)-R | TGGCGTGTCTATGATGTTCAC | ||
| erm(B) | ermB-F | CATTTAACGACGAAACTGGC | [26] |
| ermB-R | GGAACATCTGTGGTATGGCG | ||
| aac(6′)-Ie-aph(2″)-Ia | aac6-aph2F | CAGAGCCTTGGGAAGATGAAG | [27] |
| aac6-aph2R | CCTCGTGTAATTCATGTTCTGGC | ||
| sulII | Sul2-F | CGGCATCGTCAACATAACC | [28] |
| Sul2-R | GTGTGCGGATGAAGTCAG | ||
| catII | CatII-F | CCTGGAACCGCAGAGAAC | [29] |
| CatII-R | CCTGCTGAAACTTTGCCA | ||
| vanA | VanAF | GGGAAAACGACAATTGC | [30] |
| VanAR | GTACAATGCGGCCGTTA | ||
| tet(B) | tet(B)-F | CCTTATCATGCCAGTCTTGC | [28] |
| tet(B)-R | GGAACATCTGTGGTATGGCG | ||
| dfrA1 | dhfrI-F | AAGAATGGAGTTATCGGGAATG | [28] |
| dhfrI-R | GGGTAAAAACTGGCCTAAAATTG | ||
| mcr-1 | CLR5-F | ATCCCATCGCGGACAATCTC | [31] |
| Gene | Positive Control | Amplicon Size (bp) |
|---|---|---|
| blaNDM-1 | Klebsiella pneumoniae (ATCC 35657) | 621 |
| mecA | Staphylococcus aureus (ATCC 43300) | 533 |
| ampC | Staphylococcus aureus (Food isolate) | 550 |
| tet(M) | Staphylococcus aureus (ATCC 6538) | 171 |
| erm(B) | Staphylococcus aureus (ATCC 25923) | 405 |
| aac(6′)-Ie-aph(2″)-Ia | Staphylococcus aureus (Food isolate) | 348 |
| sulII | Salmonella (Food isolates) | 722 |
| catII | Enterobacter. colacae (ATCC 49141) | 495 |
| vanA | Enterobacter (Clinical isolates) | 723 |
| tet(B) | Staphylococcus aureus (ATCC 25923) | 774 |
| dfrA1 | E. coli (ATCC 25922) | 391 |
| mcr-1 | Salmonella (Food isolates) | 177 |
| Metals | Molecular Weight (g/mol) | Spik Con. (µg/L, n = 6) | Recovery (%) | Limit of Quantification (µg/L) | Uncertainty (%) |
|---|---|---|---|---|---|
| Lithium | 6.941 | 5 | 88–111 | 13.4 | 19.8 |
| Beryllium | 9.012 | 60 | 101–104 | 1.2 | 6.6 |
| Chromium | 51.99 | 15 | 85–110 | 13.1 | 37.2 |
| Cobalt | 28.010 | 5 | 88–99 | 1.3 | 14.6 |
| Arsenic | 74.9 | 5 | 99–106 | 1.4 | 12.6 |
| Cadmium | 112.4 | 5 | 84–88 | 1.4 | 28.8 |
| Lead | 207.2 | 1 | 105–108 | 1.0 | 14.8 |
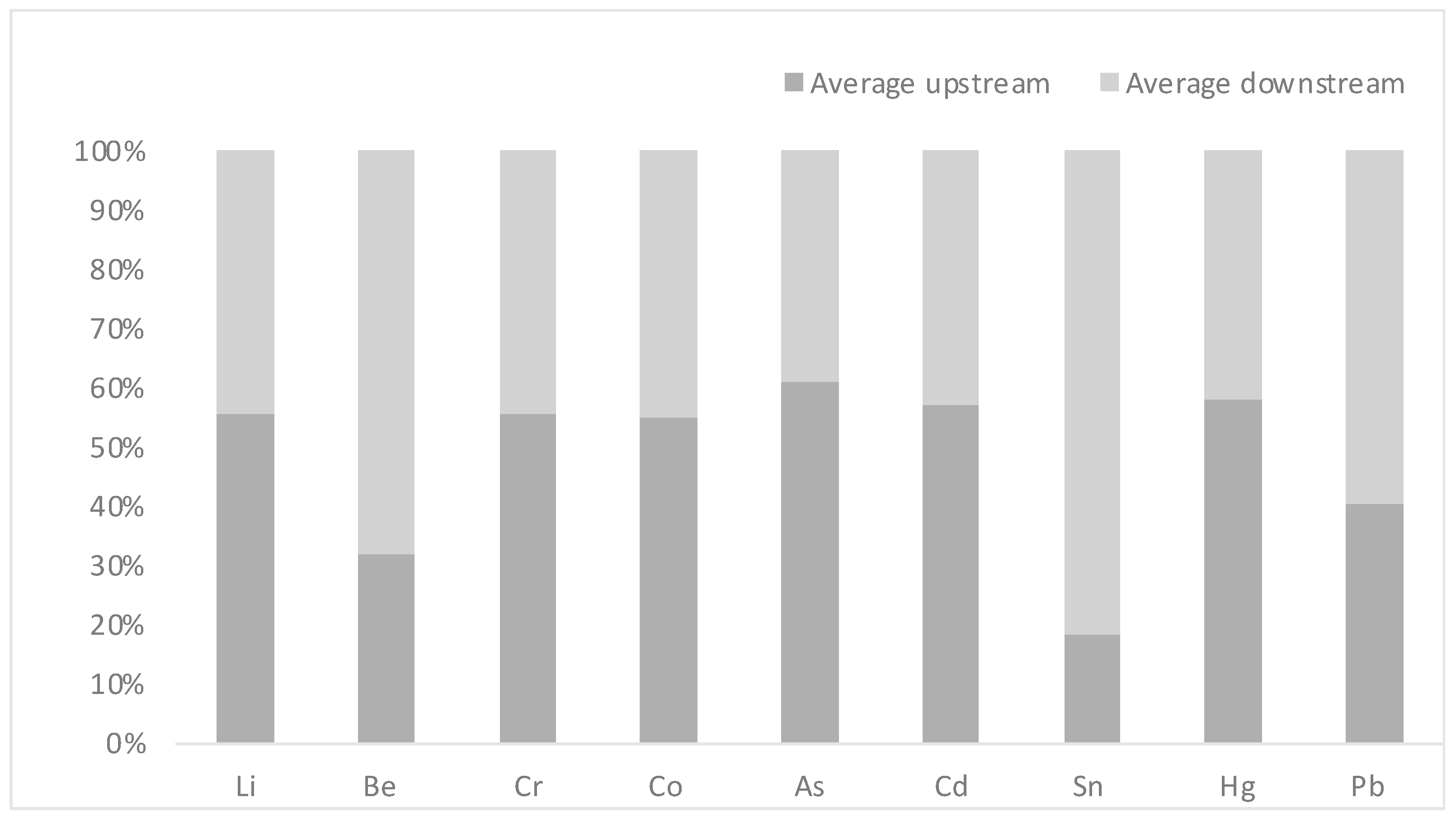
| Heavy Metal | Average Concentration Upstream (±SD) | Average Concentration Downstream (±SD) |
|---|---|---|
| Li | 40.40 (±10) | 32.32 (±2) |
| Be | 0.01 (±0.003) | 0.03 (±0.01) |
| Cr | 0.61 (±0.3) | 0.49 (±0.2) |
| Co | 0.34 (±0.06) | 0.28 (±0.1) |
| As | 1.08 (±0.46) | 0.69 (±0.1) |
| Cd | 0.01 (±0.006) | 0.01 (±0.01) |
| Sn | 65.86 (±1.1) | 296.53 (±55) |
| Hg | 0.03 (±0.01) | 0.03 (±0.004) |
| Pb | 0.20 (±0.03) | 0.30 (±0.06) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Otaibi, N.M.; Alsulaiman, B.; Alreshoodi, F.M.; Mukhtar, L.E.; Alajel, S.M.; Binsaeedan, N.M.; Alshabrmi, F.M. Screening for Antibiotic Resistance Genes in Bacteria and the Presence of Heavy Metals in the Upstream and Downstream Areas of the Wadi Hanifah Valley in Riyadh, Saudi Arabia. Antibiotics 2024, 13, 426. https://doi.org/10.3390/antibiotics13050426
Al-Otaibi NM, Alsulaiman B, Alreshoodi FM, Mukhtar LE, Alajel SM, Binsaeedan NM, Alshabrmi FM. Screening for Antibiotic Resistance Genes in Bacteria and the Presence of Heavy Metals in the Upstream and Downstream Areas of the Wadi Hanifah Valley in Riyadh, Saudi Arabia. Antibiotics. 2024; 13(5):426. https://doi.org/10.3390/antibiotics13050426
Chicago/Turabian StyleAl-Otaibi, Norah M., Bassam Alsulaiman, Fahad M. Alreshoodi, Lenah E. Mukhtar, Sulaiman M. Alajel, Norah M. Binsaeedan, and Fahad M. Alshabrmi. 2024. "Screening for Antibiotic Resistance Genes in Bacteria and the Presence of Heavy Metals in the Upstream and Downstream Areas of the Wadi Hanifah Valley in Riyadh, Saudi Arabia" Antibiotics 13, no. 5: 426. https://doi.org/10.3390/antibiotics13050426





