Molecular Mechanisms of Chlorophyll Deficiency in Ilex × attenuata ‘Sunny Foster’ Mutant
Abstract
:1. Introduction
2. Results
2.1. Yellow Hue and Brighter Color in the Mutant
2.2. Reduced Chl Synthesis in the Mutant
2.3. Varied Leaf Anatomical Structure and Impaired Chloroplast Ultrastructure in the Mutant
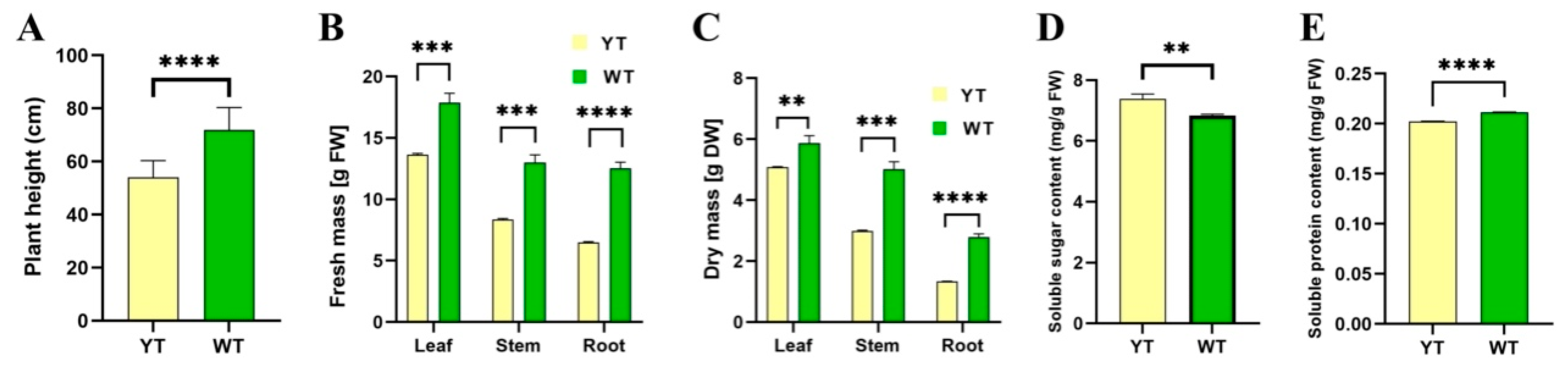
2.4. Decreased Photosynthetic Capacity and Delayed Growth in the Mutant
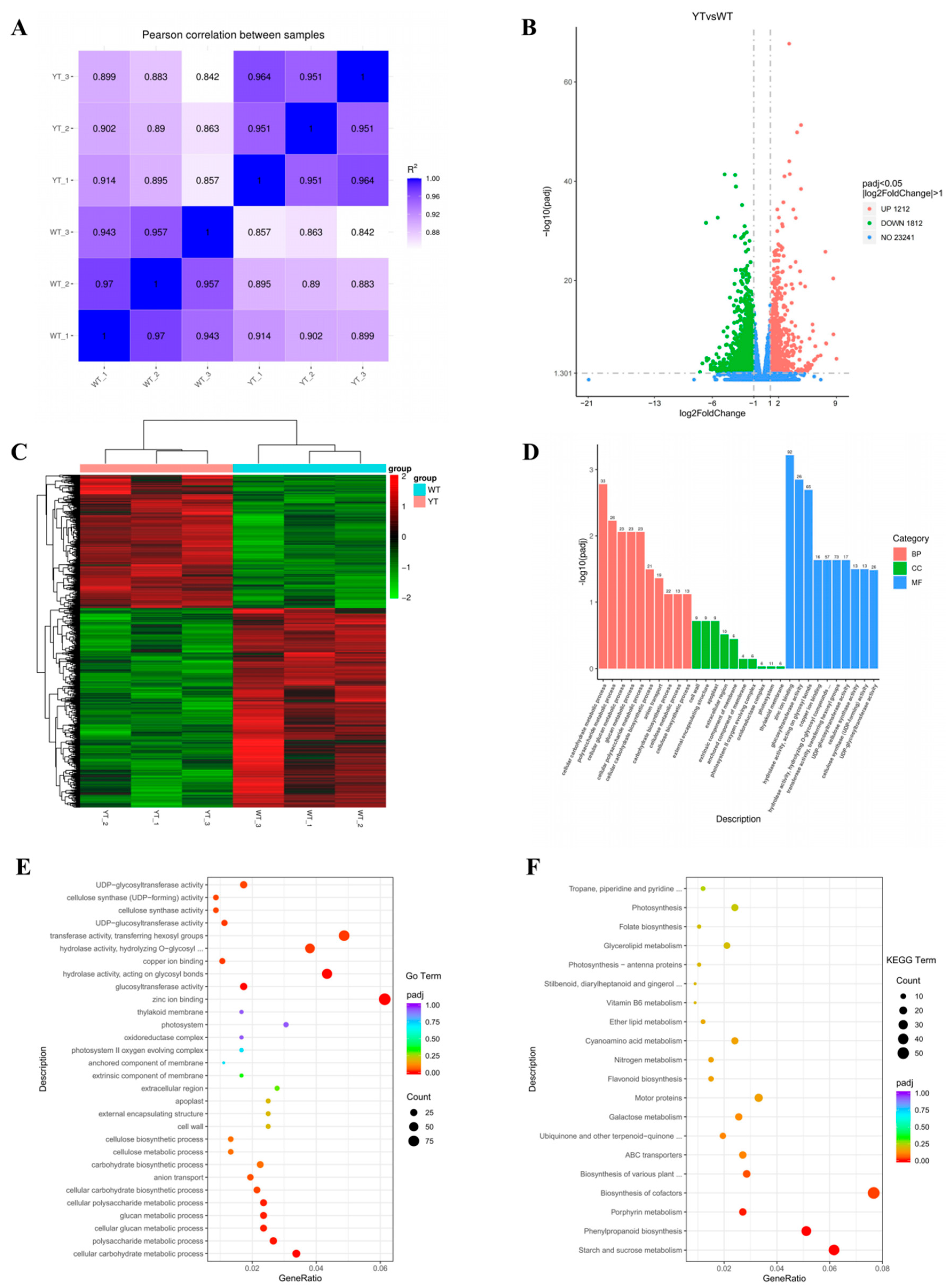
2.5. Transcriptomic Modifications in Pigment Metabolism and Photosynthesis in the Mutant
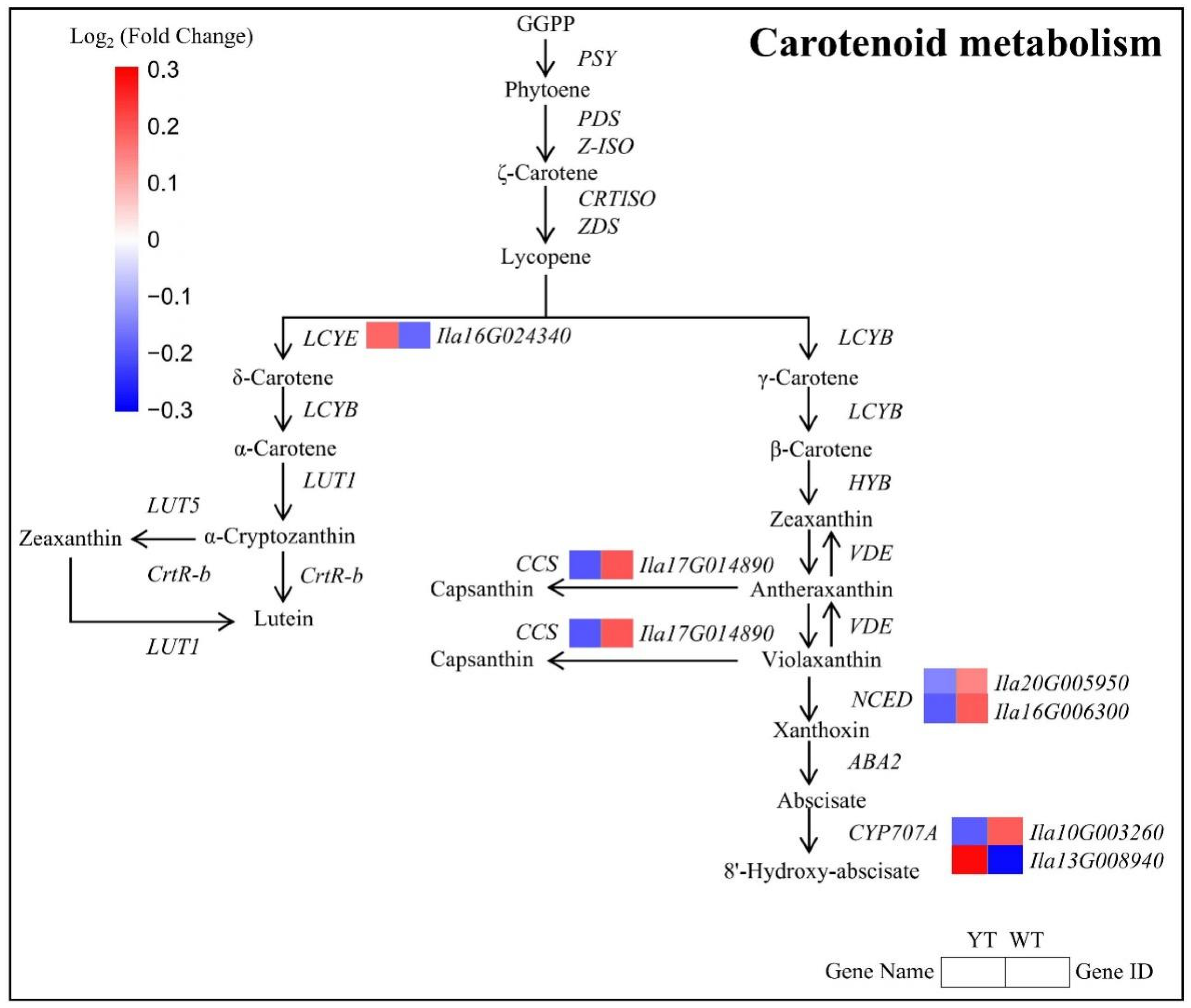
2.6. Altered Chl Biosynthesis in the Mutant
2.7. Down-Regulated Carotenoid and Flavonoid Biosynthesis in the Mutant
2.8. Down-Regulated DEGs in Photosynthesis and Chloroplast Development
2.9. Differentially Expressed Transcription Factors in the Mutant
2.10. qRT-PCR Validation of the Candidate DEGs
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Materials
5.2. Color Indice Measurement
5.3. SPAD, Pigment and Chl Precursors Content Measurements
5.4. Leaf Anatomical Structure Observation
5.5. Leaf Epidermis Microstructure Observation
5.6. Chloroplast Ultrastructure Observation
5.7. Photosynthetic Parameters Measurements
5.8. Plant Height, Biomass, Soluble Sugar, and Soluble Protein Measurements
5.9. RNA Extraction and Preparation of cDNA Library
5.10. Illumina Deep Sequencing and Data Analysis
5.11. Identification and Functional Analysis of DEGs
5.12. qRT-PCR Verification
5.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gang, H.; Li, R.; Zhao, Y.; Liu, G.; Chen, S.; Jiang, J. Loss of GLK1 Transcription Factor Function Reveals New Insights in Chlorophyll Biosynthesis and Chloroplast Development. J. Exp. Bot. 2019, 70, 3125–3138. [Google Scholar] [CrossRef] [PubMed]
- Shim, K.-C.; Kang, Y.; Song, J.-H.; Kim, Y.J.; Kim, J.K.; Kim, C.; Tai, T.H.; Park, I.; Ahn, S.-N. A Frameshift Mutation in the Mg-Chelatase I Subunit Gene OsCHLI Is Associated with a Lethal Chlorophyll-Deficient, Yellow Seedling Phenotype in Rice. Plants 2023, 12, 2831. [Google Scholar] [CrossRef] [PubMed]
- Sakowska, K.; Alberti, G.; Genesio, L.; Peressotti, A.; Delle Vedove, G.; Gianelle, D.; Colombo, R.; Rodeghiero, M.; Panigada, C.; Juszczak, R.; et al. Leaf and Canopy Photosynthesis of a Chlorophyll Deficient Soybean Mutant. Plant Cell Environ. 2018, 41, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, C.; Guzman, I.; Velasco-Cruz, C.; Bosland, P.W. Photosynthetic Pigments Profiled in Capsicum Lutescens Mutants. J. Am. Soc. Hortic. Sci. 2021, 146, 233–240. [Google Scholar] [CrossRef]
- Pao, S.-H.; Liu, J.-W.; Yang, J.-Y.; Chesson, P.; Sheue, C.-R. Uncovering the mechanisms of novel foliar variegation patterns caused by structures and pigments. Taiwania 2020, 65, 74–80. [Google Scholar] [CrossRef]
- Zhao, M.-H.; Li, X.; Zhang, X.-X.; Zhang, H.; Zhao, X.-Y. Mutation Mechanism of Leaf Color in Plants: A Review. Forests 2020, 11, 851. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, B.; Wu, J.; Li, Z.; Han, F.; Fang, Z.; Yang, L.; Zhuang, M.; Lv, H.; Liu, Y.; et al. Pigment Variation and Transcriptional Response of the Pigment Synthesis Pathway in the S2309 Triple-Color Ornamental Kale (Brassica oleracea L. var. acephala) Line. Genomics 2020, 112, 2658–2665. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zeng, F.; Song, P.; Sun, B.; Wang, Q.; Wang, J. Effects of Reduced Chlorophyll Content on Photosystem Functions and Photosynthetic Electron Transport Rate in Rice Leaves. J. Plant Physiol. 2022, 272, 153669. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, Z.; Shen, W.; Zhou, H.; Li, H.; He, X.; Li, R.; Qin, B. Disruption of the Rice ALS1 Localized in Chloroplast Causes Seedling-Lethal Albino Phenotype. Plant Sci. 2024, 338, 111925. [Google Scholar] [CrossRef]
- Yang, S.; Wang, X.; Yan, W.; Zhang, Y.; Song, P.; Guo, Y.; Xie, K.; Hu, J.; Hou, J.; Wu, Y.; et al. Melon Yellow-Green Plant (Cmygp) Encodes a Golden2-like Transcription Factor Regulating Chlorophyll Synthesis and Chloroplast Development. Theor. Appl. Genet. 2023, 136, 66. [Google Scholar] [CrossRef]
- Wang, W.; Cao, Y.; Sheng, K.; Chen, J.; Zhu, S.; Zhao, T. GhFP Positively Regulates Chlorophyll Content and Seedling Biomass in Upland Cotton. Ind. Crops Prod. 2023, 204, 117388. [Google Scholar] [CrossRef]
- Reinbothe, S.; Reinbothe, C. Regulation of Chlorophyll Biosynthesis in Angiosperms. Plant Physiol. 1996, 111, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, J.; Chen, L.; Meng, X.; Zhen, X.; Liang, Y.; Han, Y.; Li, H.; Zhang, B. Identification and Function Analysis of Yellow-Leaf Mutant (YX-Yl) of Broomcorn Millet. BMC Plant Biol. 2022, 22, 463. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Gao, Y.; Zhou, Q.; Ping, X.; Li, J.; Liu, L.; Yin, J. Genetic Mapping and Physiological Analysis of Chlorophyll-Deficient Mutant in Brassica napus L. BMC Plant Biol. 2022, 22, 244. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, S.; Lu, Z.; He, Z.; Ye, Y.; Zhao, B.; Wang, L.; Jin, B. Cytological, Physiological, and Transcriptomic Analyses of Golden Leaf Coloration in Ginkgo biloba L. Hortic. Res. 2018, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Brzezowski, P.; Richter, A.S.; Grimm, B. Regulation and Function of Tetrapyrrole Biosynthesis in Plants and Algae. Biochim. Biophys. Acta BBA Bioenerg. 2015, 1847, 968–985. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Hu, F.; Yuan, S.; Liu, C. The Identification of Key Candidate Genes Mediating Yellow Seedling Lethality in a Lilium Regale Mutant. Mol. Biol. Rep. 2020, 47, 2487–2499. [Google Scholar] [CrossRef]
- Zhang, H.; Li, J.; Yoo, J.-H.; Yoo, S.-C.; Cho, S.-H.; Koh, H.-J.; Seo, H.S.; Paek, N.-C. Rice Chlorina-1 and Chlorina-9 Encode ChlD and ChlI Subunits of Mg-Chelatase, a Key Enzyme for Chlorophyll Synthesis and Chloroplast Development. Plant Mol. Biol. 2006, 62, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, H.; Wei, L.; Guo, R.; Liu, X.; Zhang, M.; Fan, J.; Liu, S.; Liao, J.; Huang, Y.; et al. Yellow-Green Leaf 19 Encoding a Specific and Conservative Protein for Photosynthetic Organisms Affects Tetrapyrrole Biosynthesis, Photosynthesis, and Reactive Oxygen Species Metabolism in Rice. Int. J. Mol. Sci. 2023, 24, 16762. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Y.; Mazumder, A.R.; Pan, R.; Akhter, D. Research Progresses on Rice Leaf Color Mutants. Crop Des. 2022, 1, 100015. [Google Scholar] [CrossRef]
- Gao, M.; Hu, L.; Li, Y.; Weng, Y. The Chlorophyll-Deficient Golden Leaf Mutation in Cucumber Is Due to a Single Nucleotide Substitution in CsChlI for Magnesium Chelatase I Subunit. Theor. Appl. Genet. 2016, 129, 1961–1973. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, J.; Zhou, P.; Hao, M.; Zhang, M. Cytological and Transcriptomic Analysis Provide Insights into the Formation of Variegated Leaves in Ilex × Altaclerensis ‘Belgica Aurea’. Plants 2021, 10, 552. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Liu, B.; Cao, Z.; Chen, L.; Liang, Z.; Wang, M.; Liu, W.; Lin, Y.; Jiang, B. Cytological, Genetic and Transcriptomic Characterization of a Cucumber Albino Mutant. Front. Plant Sci. 2022, 13, 1047090. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Huang, Y.; Zhang, D.; Chen, H.; Liang, Y.; Hao, M.; Yin, Y. Molecular Insights into a Non-Lethal Yellow Bud Mutant in Ilex × ‘Nellie R. Stevens’. Sci. Hortic. 2024, 329, 113033. [Google Scholar] [CrossRef]
- Miura, E.; Kato, Y.; Matsushima, R.; Albrecht, V.; Laalami, S.; Sakamoto, W. The Balance between Protein Synthesis and Degradation in Chloroplasts Determines Leaf Variegation in Arabidopsis Yellow Variegated Mutants. Plant Cell 2007, 19, 1313–1328. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Zhang, C.; Gu, Y.; Cheng, R.; Huang, D.; Liu, X.; Sun, Y. Physiological and Transcriptomic Analysis of a Yellow Leaf Mutant in Watermelon. Sci. Rep. 2023, 13, 9647. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Huang, Z.; Zhao, W.; Lin, N.; Wang, Y.; Shang, F. Mechanisms for Leaf Color Changes in Osmanthus Fragrans ‘Ziyan Gongzhu’ Using Physiology, Transcriptomics and Metabolomics. BMC Plant Biol. 2023, 23, 453. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Bai, P.-P.; Gu, K.-J.; Yang, S.-Z.; Lin, H.-Y.; Shi, C.-G.; Zhao, Y.-P. Dynamic Transcriptome and Network-Based Analysis of Yellow Leaf Mutant Ginkgo biloba. BMC Plant Biol. 2022, 22, 465. [Google Scholar] [CrossRef]
- Han, H.; Zhou, Y.; Liu, H.; Chen, X.; Wang, Q.; Zhuang, H.; Sun, X.; Ling, Q.; Zhang, H.; Wang, B.; et al. Transcriptomics and Metabolomics Analysis Provides Insight into Leaf Color and Photosynthesis Variation of the Yellow-Green Leaf Mutant of Hami Melon (Cucumis melo L.). Plants 2023, 12, 1623. [Google Scholar] [CrossRef]
- Wang, F.; Chen, N.; Shen, S. iTRAQ-Based Quantitative Proteomics Analysis Reveals the Mechanism of Golden-Yellow Leaf Mutant in Hybrid Paper Mulberry. Int. J. Mol. Sci. 2022, 23, 127. [Google Scholar] [CrossRef]
- Liu, L.; Lin, N.; Liu, X.; Yang, S.; Wang, W.; Wan, X. From Chloroplast Biogenesis to Chlorophyll Accumulation: The Interplay of Light and Hormones on Gene Expression in Camellia Sinensis Cv. Shuchazao Leaves. Front. Plant Sci. 2020, 11, 256. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-G.; Xu, H.; Zhang, J.-Y.; Liang, G.-W.; Liu, Y.-T.; Guo, A.-G. Effect of Low Temperature on Chlorophyll Biosynthesis in Albinism Line of Wheat (Triticum aestivum) FA85. Physiol. Plant. 2012, 145, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Yangen, F.; Xiuxiu, Z.; Hanyue, W.; Yueyue, T.; Qinzeng, X.; Lixia, Z. Effects of Light Intensity on Metabolism of Light-Harvesting Pigment and Photosynthetic System in Camellia sinensis L. Cultivar ‘Huangjinya’. Environ. Exp. Bot. 2019, 166, 103796. [Google Scholar] [CrossRef]
- Dong, H.; Fei, G.-L.; Wu, C.-Y.; Wu, F.-Q.; Sun, Y.-Y.; Chen, M.-J.; Ren, Y.-L.; Zhou, K.-N.; Cheng, Z.-J.; Wang, J.-L.; et al. A Rice Virescent-Yellow Leaf Mutant Reveals New Insights into the Role and Assembly of Plastid Caseinolytic Protease in Higher Plants. Plant Physiol. 2013, 162, 1867–1880. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, Y.; Niu, Y.; Zhang, X.; Zhao, J.; Sun, L.; Wang, H.; Xiao, J.; Wang, X. Characterization and Fine Mapping of a Leaf Yellowing Mutant in Common Wheat. Plant Growth Regul. 2020, 92, 233–247. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, T.; Fu, F.-F.; El-Kassaby, Y.A.; Wang, G. Metabolome and Transcriptome Analyses Reveal the Regulatory Mechanisms of Photosynthesis in Developing Ginkgo biloba Leaves. Int. J. Mol. Sci. 2021, 22, 2601. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Huang, L.; Chen, Q.; Lv, Y.; Sun, H.; Liang, Z. Physiological and Anatomical Differences and Differentially Expressed Genes Reveal Yellow Leaf Coloration in Shumard Oak. Plants 2020, 9, 169. [Google Scholar] [CrossRef]
- Zuo, L.; Zhang, S.; Liu, Y.; Huang, Y.; Yang, M.; Wang, J. The Reason for Growth Inhibition of Ulmus pumila ‘Jinye’: Lower Resistance and Abnormal Development of Chloroplasts Slow Down the Accumulation of Energy. Int. J. Mol. Sci. 2019, 20, 4227. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Wang, J.; Wang, Q.; Zhang, G.; Zhao, Y.; Ma, Q.; Wu, Z.; Ma, J.; Gu, C. Decoding the Formation of Diverse Petal Colors of Lagerstroemia indica by Integrating the Data from Transcriptome and Metabolome. Front. Plant Sci. 2022, 13, 970023. [Google Scholar] [CrossRef]
- Zhu, L.; Wen, J.; Ma, Q.; Yan, K.; Du, Y.; Chen, Z.; Lu, X.; Ren, J.; Wang, Y.; Li, S.; et al. Transcriptome Profiling Provides Insights into Leaf Color Changes in Two Acer palmatum Genotypes. BMC Plant Biol. 2022, 22, 589. [Google Scholar] [CrossRef]
- Yao, X.; Zhang, F.; Corlett, R.T. Utilization of the Hollies (Ilex L. spp.): A Review. Forests 2022, 13, 94. [Google Scholar] [CrossRef]
- Bailes, C.; Andrews, S. Hollies for Gardeners; Timber Press: Portland, OR, USA, 2006. [Google Scholar]
- Galle, F.C. Hollies: The Genus Ilex; Timber Press: Portland, OR, USA, 1997. [Google Scholar]
- Beyond Galle: A Compilation of Cultivated Ilex Not Included in Fred Galle’s “Hollies: The Genus Ilex”. Available online: https://www.hollysocam.org/untitled/memberindex/PDF/Beyond-Galle.pdf (accessed on 10 December 2023).
- Holly of the Year 2006. Available online: https://www.hollysocam.org/holly-year-6.htm (accessed on 25 December 2023).
- Application of Ilex Plants in Urban Greening. Available online: https://www.proquest.com/openview/eb1962677c6ccfe6e2efcede33267740/1?pq-origsite=gscholar&cbl=1596366 (accessed on 2 March 2024).
- Veil, J.M. Ilex at the University of Delaware Botanic Gardens: A Template for Measuring Collection Relevance at Small University Gardens. Ph.D. Thesis, University of Delaware, Newark, DE, USA, 2015. [Google Scholar]
- Chang, Q.S.; Zhang, L.X.; Hou, X.G.; Wang, Z.; Wang, N.; Gong, M.G.; Zhang, Q.M.; Chen, H.; Shi, Z.Q.; Deng, C.C. The Anatomical, Physiological, and Molecular Analysis of a Chlorophyll-Deficient Mutant in Tree Peony (Paeonia suffruticosa). Photosynthetica 2019, 57, 724–730. [Google Scholar] [CrossRef]
- Gan, Y.; Kou, Y.; Yan, F.; Wang, X.; Wang, H.; Song, X.; Zhang, M.; Zhao, X.; Jia, R.; Ge, H.; et al. Comparative Transcriptome Profiling Analysis Reveals the Adaptive Molecular Mechanism of Yellow-Green Leaf in Rosa Beggeriana “Aurea”. Front. Plant Sci. 2022, 13, 845662. [Google Scholar] [CrossRef] [PubMed]
- Rigon, J.P.G.; Capuani, S.; Fernandes, D.M.; Guimarães, T.M. A Novel Method for the Estimation of Soybean Chlorophyll Content Using a Smartphone and Image Analysis. Photosynthetica 2016, 54, 559–566. [Google Scholar] [CrossRef]
- Falbel, T.G.; Staehelin, L.A. Partial Blocks in the Early Steps of the Chlorophyll Synthesis Pathway: A Common Feature of Chlorophyll b-Deficient Mutants. Physiol. Plant. 1996, 97, 311–320. [Google Scholar] [CrossRef]
- Li, S.; Wang, S.; Wang, P.; Gao, L.; Yang, R.; Li, Y. Label-Free Comparative Proteomic and Physiological Analysis Provides Insight into Leaf Color Variation of the Golden-Yellow Leaf Mutant of Lagerstroemia indica. J. Proteom. 2020, 228, 103942. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Grimm, B. Connecting Chlorophyll Metabolism with Accumulation of the Photosynthetic Apparatus. Trends Plant Sci. 2021, 26, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Chen, M. Chlorophyll Modifications and Their Spectral Extension in Oxygenic Photosynthesis. Annu. Rev. Biochem. 2014, 83, 317–340. [Google Scholar] [CrossRef] [PubMed]
- Rogowski, P.; Wasilewska-Dębowska, W.; Krupnik, T.; Drożak, A.; Zienkiewicz, M.; Krysiak, M.; Romanowska, E. Photosynthesis and Organization of Maize Mesophyll and Bundle Sheath Thylakoids of Plants Grown in Various Light Intensities. Environ. Exp. Bot. 2019, 162, 72–86. [Google Scholar] [CrossRef]
- Schmidt, S.B.; Eisenhut, M.; Schneider, A. Chloroplast Transition Metal Regulation for Efficient Photosynthesis. Trends Plant Sci. 2020, 25, 817–828. [Google Scholar] [CrossRef]
- Feng, Y.; Li, Y.; Zhao, Y.; Shi, H. Physiological, Transcriptome and Co-Expression Network Analysis of Chlorophyll-Deficient Mutants in Flue-Cured Tobacco. BMC Plant Biol. 2023, 23, 153. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Zhang, W.; Liu, Y.; Zhang, H.; Ren, H.; Chen, Y.; Wang, L.; Zeng, J.; Yang, Y.; Wang, X. Pale Green Mutant Analyses Reveal the Importance of CsGLKs in Chloroplast Developmental Regulation and Their Effects on Flavonoid Biosynthesis in Tea Plant. Plant Physiol. Biochem. 2020, 146, 392–402. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, M.; Ruan, J. Integrated Transcriptome and Metabolic Analyses Reveals Novel Insights into Free Amino Acid Metabolism in Huangjinya Tea Cultivar. Front. Plant Sci. 2017, 8, 291. [Google Scholar] [CrossRef]
- Dong, S.; Fan, M.; Qin, Q.; Zhang, Z.; Duan, K.; Ćosić, T.; Raspor, M.; Ni, D. Natural Albino Mutant of Daylily (Hemerocallis spp.) Reveals a Link between Drought Sensitivity and Photosynthetic Pigments Metabolism. Front. Biosci. 2024, 29, 60. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; He, D.; Bai, M.; Li, B.; Zhang, Q.; Luo, L. Cytological, Physiological and Transcriptomic Analysis of Variegated Leaves in Primulina pungentisepala Offspring. BMC Plant Biol. 2022, 22, 419. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, Z.; Zhang, M.; Wang, J.; Cheng, T.; Zhang, Q.; Pan, H. FsHemF Is Involved in the Formation of Yellow Forsythia Leaves by Regulating Chlorophyll Synthesis in Response to Light Intensity. Plant Physiol. Biochem. 2023, 200, 107746. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Li, G.; Liu, N.; Li, Y.; Li, J.; Huang, X.; Zhao, D. Identification and Characterization of a Novel Yellow Leaf Mutant Yl1 in Rice. Phyton Int. J. Exp. Bot. 2022, 91, 2419–2437. [Google Scholar] [CrossRef]
- Beale, S.I. Green Genes Gleaned. Trends Plant Sci. 2005, 10, 309–312. [Google Scholar] [CrossRef]
- Zeng, Z.; Lin, T.; Zhao, J.; Zheng, T.; Xu, L.; Wang, Y.; Liu, L.; Jiang, L.; Chen, S.; Wan, J. OsHemA Gene, Encoding Glutamyl-tRNA Reductase (GluTR) Is Essential for Chlorophyll Biosynthesis in Rice (Oryza Sativa). J. Integr. Agric. 2020, 19, 612–623. [Google Scholar] [CrossRef]
- Tanaka, R.; Tanaka, A. Tetrapyrrole Biosynthesis in Higher Plants. Annu. Rev. Plant Biol. 2007, 58, 321–346. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, X.; Zhang, Y.; Zhou, F.; Zhu, P. Simultaneous Changes in Anthocyanin, Chlorophyll, and Carotenoid Contents Produce Green Variegation in Pink–Leaved Ornamental Kale. BMC Genom. 2021, 22, 455. [Google Scholar] [CrossRef] [PubMed]
- Willows, R.D. Biosynthesis of Chlorophylls from Protoporphyrin IX. Nat. Prod. Rep. 2003, 20, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Al-Karadaghi, S.; Franco, R.; Hansson, M.; Shelnutt, J.A.; Isaya, G.; Ferreira, G.C. Chelatases: Distort to Select? Trends Biochem. Sci. 2006, 31, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-Y.; Shi, J.-C.; Wang, D.-J.; Liang, X.; Wei, F.; Gong, C.-M.; Qiu, L.-J.; Zhou, H.-C.; Folta, K.M.; Wen, Y.-Q.; et al. A Point Mutation in the Gene Encoding Magnesium Chelatase I Subunit Influences Strawberry Leaf Color and Metabolism. Plant Physiol. 2023, 192, 2737–2755. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Hou, Y.; Zheng, L.; Shi, H.; Liu, Z.; Wang, Y.; Li, S.; Liu, L.; Guo, M.; Yang, Z.; et al. Characterization and Fine Mapping of a Yellow Leaf Gene Regulating Chlorophyll Biosynthesis and Chloroplast Development in Cotton (Gossypium arboreum). Gene 2023, 885, 147712. [Google Scholar] [CrossRef] [PubMed]
- Terry, M.J.; Kendrick, R.E. Feedback Inhibition of Chlorophyll Synthesis in the Phytochrome Chromophore-Deficient Aurea Andyellow-Green-2 Mutants of Tomato. Plant Physiol. 1999, 119, 143–152. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Chai, J.; Wang, L.; Wang, C.; Long, W.; Wang, D.; Wang, Y.; Zheng, M.; Peng, C.; et al. Green-Revertible Chlorina 1 (Grc1) Is Required for the Biosynthesis of Chlorophyll and the Early Development of Chloroplasts in Rice. J. Plant Biol. 2013, 56, 326–335. [Google Scholar] [CrossRef]
- Yang, M.; Wan, S.; Chen, J.; Chen, W.; Wang, Y.; Li, W.; Wang, M.; Guan, R. Mutation to a Cytochrome P450-like Gene Alters the Leaf Color by Affecting the Heme and Chlorophyll Biosynthesis Pathways in Brassica napus. Plant J. 2023, 116, 432–445. [Google Scholar] [CrossRef]
- Sato, T.; Shimoda, Y.; Matsuda, K.; Tanaka, A.; Ito, H. Mg-Dechelation of Chlorophyll a by Stay-Green Activates Chlorophyll b Degradation through Expressing Non-Yellow Coloring 1 in Arabidopsis thaliana. J. Plant Physiol. 2018, 222, 94–102. [Google Scholar] [CrossRef]
- Park, S.-Y.; Yu, J.-W.; Park, J.-S.; Li, J.; Yoo, S.-C.; Lee, N.-Y.; Lee, S.-K.; Jeong, S.-W.; Seo, H.S.; Koh, H.-J.; et al. The Senescence-Induced Staygreen Protein Regulates Chlorophyll Degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of Plant Pigments: Anthocyanins, Betalains and Carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Kerfeld, C.A. Water-Soluble Carotenoid Proteins of Cyanobacteria. Arch. Biochem. Biophys. 2004, 430, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, X.; Xie, J.; Ding, W.; Li, Y.; Yue, Y.; Wang, L. Biochemical and Comparative Transcriptome Analyses Reveal Key Genes Involved in Major Metabolic Regulation Related to Colored Leaf Formation in Osmanthus Fragrans ‘Yinbi Shuanghui’ during Development. Biomolecules 2020, 10, 549. [Google Scholar] [CrossRef]
- Breitholtz, H.-L.; Srivastava, R.; Tyystjärvi, E.; Rintamäki, E. LHC II Protein Phosphorylation in Leaves of Arabidopsis thaliana Mutants Deficient in Non-Photochemical Quenching. Photosynth. Res. 2005, 84, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Llorente, B.; Torres-Montilla, S.; Morelli, L.; Florez-Sarasa, I.; Matus, J.T.; Ezquerro, M.; D’Andrea, L.; Houhou, F.; Majer, E.; Picó, B.; et al. Synthetic Conversion of Leaf Chloroplasts into Carotenoid-Rich Plastids Reveals Mechanistic Basis of Natural Chromoplast Development. Proc. Natl. Acad. Sci. USA 2020, 117, 21796–21803. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Blankenship, R.E. On the Interface of Light-Harvesting Antenna Complexes and Reaction Centers in Oxygenic Photosynthesis. Biochim. Biophys. Acta BBA Bioenerg. 2019, 1860, 148079. [Google Scholar] [CrossRef]
- Gao, P.; Xia, H.; Li, Q.; Li, Z.; Zhai, C.; Weng, L.; Mi, H.; Yan, S.; Datla, R.; Wang, H.; et al. PALE-GREEN LEAF 1, a Rice cpSRP54 Protein, Is Essential for the Assembly of the PSI-LHCI Supercomplex. Plant Direct 2022, 6, e436. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.-X.; Zhang, R.-X.; Liu, S.; He, Y.-M.; Feng, X.-H.; Ul Haq, S.; Luo, D.-X.; Gong, Z.-H. Leaf-Color Mutation Induced by Ethyl Methane Sulfonate and Genetic and Physio-Biochemical Characterization of Leaf-Color Mutants in Pepper (Capsicum annuum L.). Sci. Hortic. 2019, 257, 108709. [Google Scholar] [CrossRef]
- Mao, L.; Dai, Y.; Huang, Y.; Yang, S.; Sun, H.; Zhou, Y.; Sun, Y.; Yang, B.; Zou, X.; Liu, Z. Studying the Effect of Light Intensity on the Photosynthetic Mechanism of Pepper Leaf Yellowing Mutants by Proteomics and Phosphoproteomics. Plant Sci. 2023, 334, 111763. [Google Scholar] [CrossRef]
- Jenny, A.; Mark, W.; Robin, G.W.; Caroline, A.H.; Alexander, V.R.; Peter, H.; Stefan, J. Absence of the Lhcb1 and Lhcb2 Proteins of the Light-Harvesting Complex of Photosystem II—Effects on Photosynthesis, Grana Stacking and Fitness. Plant J. 2003, 35, 350–361. [Google Scholar] [CrossRef]
- Koyama, T. Regulatory Mechanisms of Transcription Factors in Plant Morphology and Function. Int. J. Mol. Sci. 2023, 24, 7039. [Google Scholar] [CrossRef] [PubMed]
- Niu, F.; Cui, X.; Zhao, P.; Sun, M.; Yang, B.; Deyholos, M.K.; Li, Y.; Zhao, X.; Jiang, Y.-Q. WRKY42 Transcription Factor Positively Regulates Leaf Senescence through Modulating SA and ROS Synthesis in Arabidopsis thaliana. Plant J. 2020, 104, 171–184. [Google Scholar] [CrossRef]
- Piao, W.; Kim, S.-H.; Lee, B.-D.; An, G.; Sakuraba, Y.; Paek, N.-C. Rice Transcription Factor OsMYB102 Delays Leaf Senescence by Down-Regulating Abscisic Acid Accumulation and Signaling. J. Exp. Bot. 2019, 70, 2699–2715. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mao, L.; Yang, B.; Cui, Q.; Dai, Y.; Li, X.; Chen, Y.; Dai, X.; Zou, X.; Ou, L.; et al. A Multi-Omics Approach Identifies bHLH71-like as a Positive Regulator of Yellowing Leaf Pepper Mutants Exposed to High-Intensity Light. Hortic. Res. 2023, 10, uhad098. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Chen, L.; Yu, Q.; Zhou, W.; Gou, X.; Li, J.; Hou, S. Multiple Transcriptional Factors Control Stomata Development in Rice. New Phytol. 2019, 223, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Oda-Yamamizo, C.; Mitsuda, N.; Sakamoto, S.; Ogawa, D.; Ohme-Takagi, M.; Ohmiya, A. The NAC Transcription Factor ANAC046 Is a Positive Regulator of Chlorophyll Degradation and Senescence in Arabidopsis Leaves. Sci. Rep. 2016, 6, 23609. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shi, D.; Wang, Y.; Zhang, L.; Chen, X.; Yang, X.; Xiong, H.; Bhattarai, G.; Ravelombola, W.; Olaoye, D.; et al. Transcript Profiling for Regulation of Sweet Potato Skin Color in Sushu8 and Its Mutant Zhengshu20. Plant Physiol. Biochem. 2020, 148, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liang, D.; Xu, Q.; Yang, F.; Zhu, G. Involvement of CsERF2 in Leaf Variegation of Cymbidium Sinense ‘Dharma’. Planta 2020, 252, 29. [Google Scholar] [CrossRef]
- Liu, N.-T.; Jane, W.-N.; Tsay, H.-S.; Wu, H.; Chang, W.-C.; Lin, C.-S. Chloroplast Genome Aberration in Micropropagation-Derived Albino Bambusa Edulis Mutants, Ab1 and Ab2. Plant Cell Tissue Organ Cult. 2007, 88, 147–156. [Google Scholar] [CrossRef]
- Moon, J.; Zhu, L.; Shen, H.; Huq, E. PIF1 Directly and Indirectly Regulates Chlorophyll Biosynthesis to Optimize the Greening Process in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 9433–9438. [Google Scholar] [CrossRef]
- Chen, M.; Ji, M.; Wen, B.; Liu, L.; Li, S.; Chen, X.; Gao, D.; Li, L. GOLDEN 2-LIKE Transcription Factors of Plants. Front. Plant Sci. 2016, 7, 1509. [Google Scholar] [CrossRef] [PubMed]
- Fitter, D.W.; Martin, D.J.; Copley, M.J.; Scotland, R.W.; Langdale, J.A. GLK Gene Pairs Regulate Chloroplast Development in Diverse Plant Species. Plant J. 2002, 31, 713–727. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Muramatsu, M.; Hakata, M.; Ueno, O.; Nagamura, Y.; Hirochika, H.; Takano, M.; Ichikawa, H. Ectopic Overexpression of The Transcription Factor OsGLK1 Induces Chloroplast Development in Non-Green Rice Cells. Plant Cell Physiol. 2009, 50, 1933–1949. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Wang, P.; Korkaric, M.; Capper, R.G.; Saunders, N.J.; Langdale, J.A. GLK Transcription Factors Coordinate Expression of the Photosynthetic Apparatus in Arabidopsis. Plant Cell 2009, 21, 1109–1128. [Google Scholar] [CrossRef] [PubMed]
- An, X.-H.; Tian, Y.; Chen, Y.-H.; Li, E.-M.; Li, M.; Cheng, C.-G. Functional Identification of Apple MdGLK1 Which Regulates Chlorophyll Biosynthesis in Arabidopsis. J. Plant Growth Regul. 2019, 38, 778–787. [Google Scholar] [CrossRef]
- Gonnet, J.-F. Colour Effects of Co-Pigmentation of Anthocyanin Revisited—3. A Further Description Using CIELAB Differences and Assessment of Matched Colours Using the CMC Model. Food Chem. 2001, 75, 473–485. [Google Scholar] [CrossRef]
- Wrolstad, R.E.; Durst, R.W.; Lee, J. Tracking Color and Pigment Changes in Anthocyanin Products. Trends Food Sci. Technol. 2005, 16, 423–428. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. [34] Chlorophylls and Carotenoids: Pigments of Photosynthetic Biomembranes. In Methods in Enzymology; Plant Cell Membranes; Academic Press: Cambridge, MA, USA, 1987; Volume 148, pp. 350–382. [Google Scholar] [CrossRef]
- Hughes, N.M.; Morley, C.B.; Smith, W.K. Coordination of Anthocyanin Decline and Photosynthetic Maturation in Juvenile Leaves of Three Deciduous Tree Species. New Phytol. 2007, 175, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Manetas, Y.; Petropoulou, Y.; Psaras, G.K.; Drinia, A. Exposed Red (Anthocyanic) Leaves of Quercus Coccifera Display Shade Characteristics. Funct. Plant Biol. 2003, 30, 265–270. [Google Scholar] [CrossRef]
- Ma, D.; Sun, D.; Wang, C.; Li, Y.; Guo, T. Expression of Flavonoid Biosynthesis Genes and Accumulation of Flavonoid in Wheat Leaves in Response to Drought Stress. Plant Physiol. Biochem. 2014, 80, 60–66. [Google Scholar] [CrossRef]
- Qian, P.; Marklew, C.J.; Viney, J.; Davison, P.A.; Brindley, A.A.; Söderberg, C.; Al-Karadaghi, S.; Bullough, P.A.; Grossmann, J.G.; Hunter, C.N. Structure of the Cyanobacterial Magnesium Chelatase H Subunit Determined by Single Particle Reconstruction and Small-Angle X-Ray Scattering. J. Biol. Chem. 2012, 287, 4946–4956. [Google Scholar] [CrossRef] [PubMed]
- Kumachova, T.; Babosha, A.; Ryabchenko, A.; Ivanova, T.; Voronkov, A. Leaf Epidermis in Rosaceae: Diversity of the Cuticular Folding and Microstructure. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2021, 91, 455–470. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, S.; Song, S.; Xu, F.; Pan, Y.; Wang, H. Transcriptomic and Proteomic Analyses Reveal New Insight into Chlorophyll Synthesis and Chloroplast Structure of Maize Leaves under Zinc Deficiency Stress. J. Proteom. 2019, 199, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Meravi, N.; Kumar Prajapati, S. Effect Street Light Pollution on the Photosynthetic Efficiency of Different Plants. Biol. Rhythm Res. 2020, 51, 67–75. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.; Hamilton, J.K.; Rebers, P.A.; Smith, F. A Colorimetric Method for the Determination of Sugars. Nature 1951, 168, 167. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Chong, X.; Wang, Y.; Xu, X.; Zhang, F.; Wang, C.; Zhou, Y.; Zhou, T.; Li, Y.; Lu, X.; Chen, H. Efficient Virus-Induced Gene Silencing in Ilex Dabieshanensis Using Tobacco Rattle Virus. Forests 2023, 14, 488. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]







| Ultrastructural Parameters | YT | WT |
|---|---|---|
| Chloroplast number (per cell) | 3.90 ± 0.74 b | 9.60 ± 0.84 a |
| Chloroplast length (um) | 6.46 ± 0.86 a | 5.98 ± 0.80 a |
| Chloroplast width (um) | 5.34 ± 1.16 a | 2.31 ± 0.48 b |
| Chloroplast length/width | 1.25 ± 0.29 b | 2.67 ± 0.52 a |
| Starch grain number (per cell) | 0.00 ± 0.00 b | 2.10 ± 0.57 a |
| Parameters | YT | WT | |
|---|---|---|---|
| Photosynthesis | Pn [μmol(CO2) m−2 s−1] | 0.37 ± 0.55 b | 19.47 ± 2.14 a |
| E [mol(H2O) m−2 s−1] | 0.87 ± 0.21 b | 4.33 ± 0.38 a | |
| Gs [mol(H2O) m−2 s−1] | 40.67 ± 8.33 b | 296.00 ± 43.35 a | |
| Ci [μmol(CO2) mol−1] | 428.00 ± 26.00 a | 274.67 ± 5.13 b | |
| Chl fluorescence | Fo | 153.00 ± 13.00 b | 217.33 ± 24.58 a |
| Fm | 184.33 ± 13.80 b | 762.00 ± 61.88 a | |
| Fv | 31.33 ± 4.62 b | 544.67 ± 37.63 a | |
| Fv/Fm | 0.17 ± 0.03 b | 0.72 ± 0.01 a | |
| ΦPSII | 0.11 ± 0.05 b | 0.48 ± 0.01 a | |
| NPQ | 0.02 ± 0.02 b | 0.48 ± 0.11 a | |
| Y (NO) | 0.87 ± 0.04 a | 0.35 ± 0.02 b | |
| Y (NPQ) | 0.02 ± 0.02 b | 0.17 ± 0.03 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, Y.; Huang, Y.; Zhang, D.; Chen, H.; Liang, Y.; Hao, M.; Yin, Y. Molecular Mechanisms of Chlorophyll Deficiency in Ilex × attenuata ‘Sunny Foster’ Mutant. Plants 2024, 13, 1284. https://doi.org/10.3390/plants13101284
Zou Y, Huang Y, Zhang D, Chen H, Liang Y, Hao M, Yin Y. Molecular Mechanisms of Chlorophyll Deficiency in Ilex × attenuata ‘Sunny Foster’ Mutant. Plants. 2024; 13(10):1284. https://doi.org/10.3390/plants13101284
Chicago/Turabian StyleZou, Yiping, Yajian Huang, Donglin Zhang, Hong Chen, Youwang Liang, Mingzhuo Hao, and Yunlong Yin. 2024. "Molecular Mechanisms of Chlorophyll Deficiency in Ilex × attenuata ‘Sunny Foster’ Mutant" Plants 13, no. 10: 1284. https://doi.org/10.3390/plants13101284





