Research on the Functional Microbe Activation System in a Post-Polymer Flooded Reservoir
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Collection
2.2. PCR Amplification and 16S rRNA Gene Sequencing
2.3. Sequencing Data Analysis
2.4. Activation of Indigenous Functional Microbes Test
2.5. Bacteria Growth Curve
2.6. Determination of Crude Oil Biodegradation Rate
2.7. Core Oil Displacement Experiment
3. Results
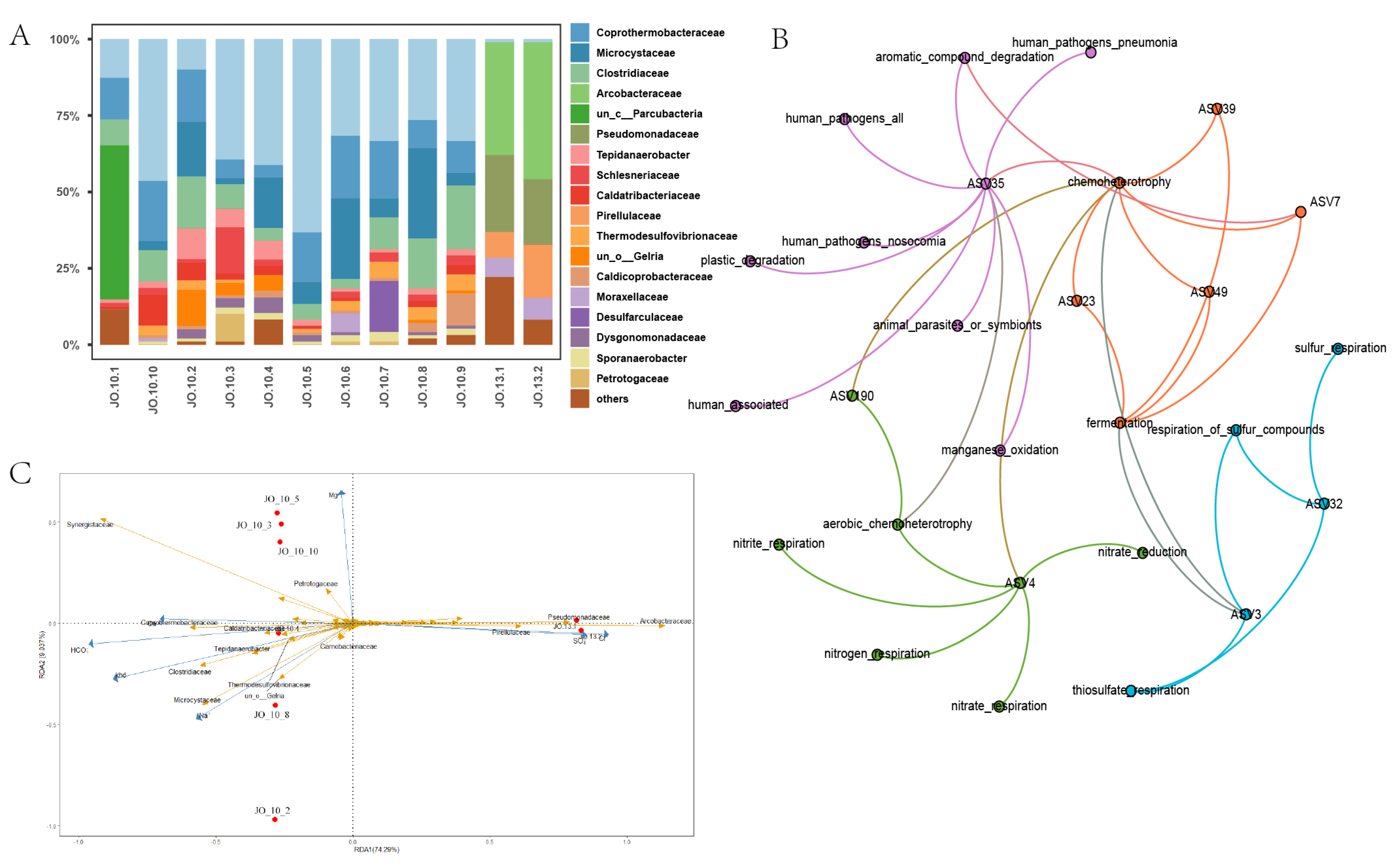
3.1. Characteristics of Microbial Community in Oil Reservoirs after Polymer Flooding
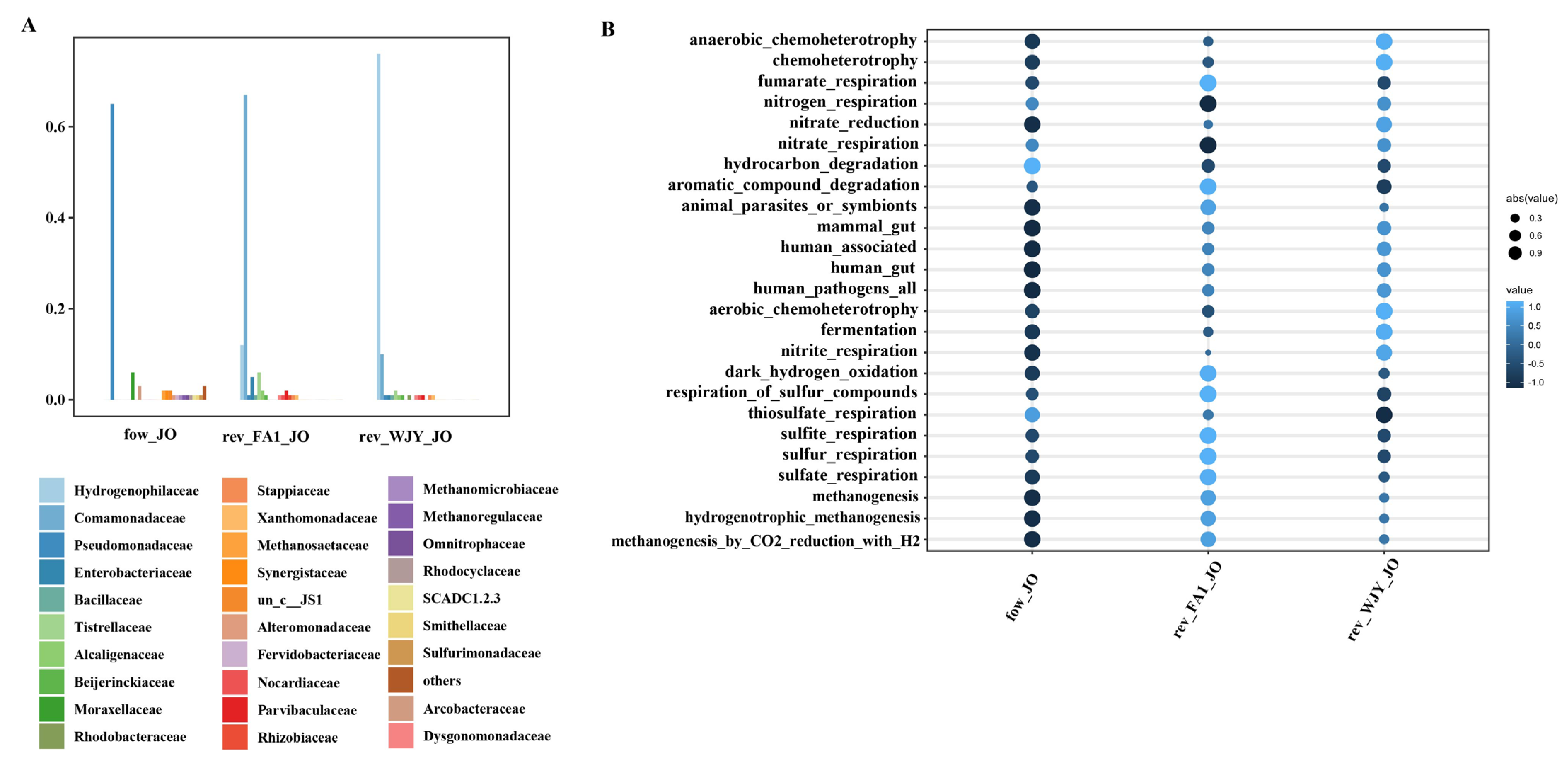
3.2. Research on Activation of Functional Microorganisms
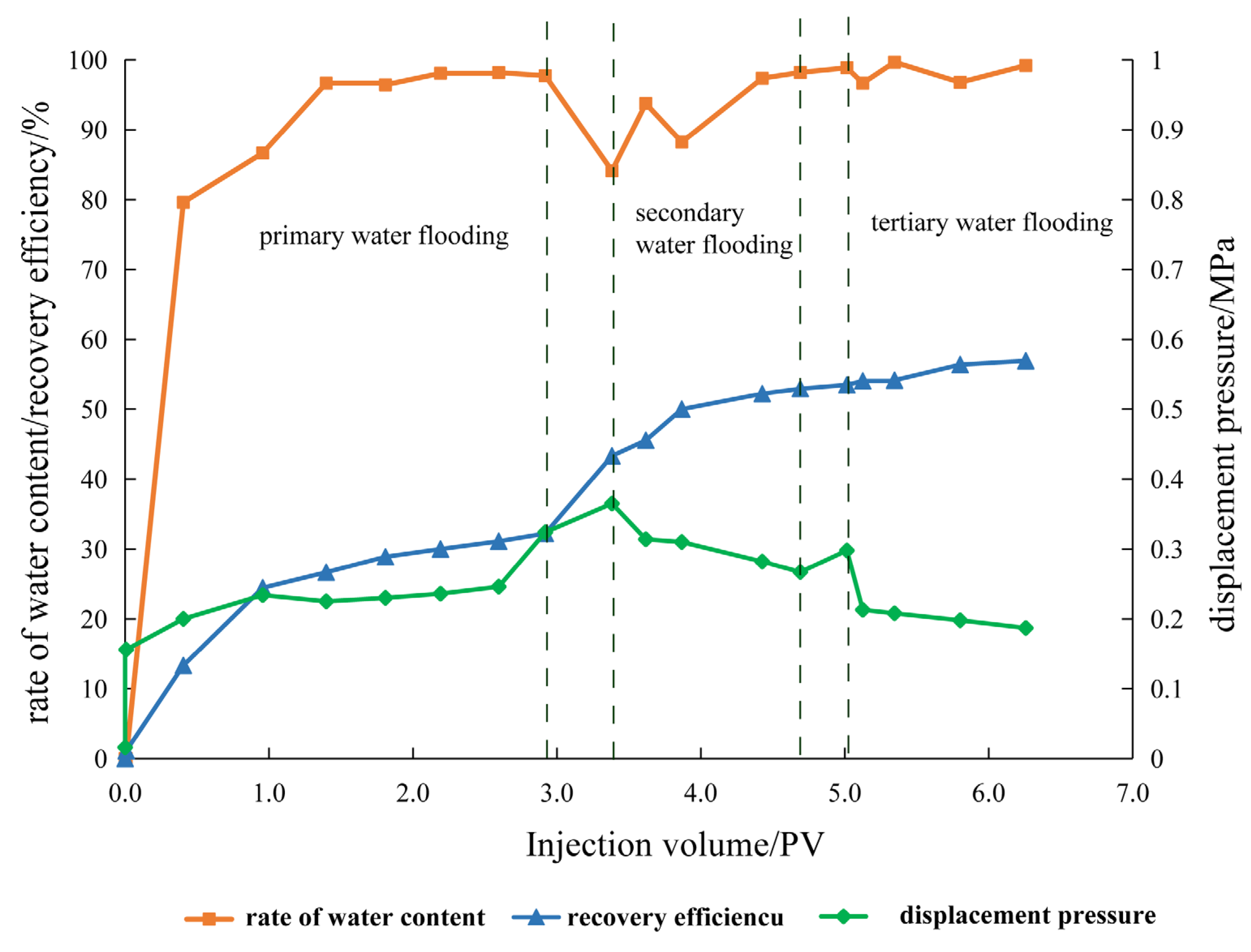
3.3. Core Displacement Experiment
3.4. Changes in Community Structure during Activation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- El-hoshoudy, A.N. Experimental and theoretical investigation for synthetic polymers, biopolymers and polymeric nanocomposites application in enhanced oil recovery operations. Arab. J. Sci. Eng. 2022, 47, 10887–10915. [Google Scholar] [CrossRef]
- Ruiz-Cañas, M.C.; Quintero-Perez, H.I.; Castro-Garcia, R.H.; Romero-Bohorquez, A.R. Use of nanoparticles to improve thermochemical resistance of synthetic polymer to enhanced oil recovery applications: A review. CTF-Cienc. Tecnol. Y Futuro 2020, 10, 85–97. [Google Scholar]
- Tavakkoli, O.; Kamyab, H.; Shariati, M.; Mohamed, A.M.; Junin, R. Effect of nanoparticles on the performance of polymer/surfactant flooding for enhanced oil recovery: A review. Fuel 2022, 312, 122867. [Google Scholar] [CrossRef]
- Firozjaii, A.M.; Saghafi, H.R. Review on chemical enhanced oil recovery using polymer flooding: Fundamentals, experimental and numerical simulation. Petroleum 2020, 6, 115–122. [Google Scholar] [CrossRef]
- Gbadamosi, A.O.; Junin, R.; Manan, M.A.; Agi, A.; Yusuff, A.S. An overview of chemical enhanced oil recovery: Recent advances and prospects. Int. Nano Lett. 2019, 9, 171–202. [Google Scholar] [CrossRef]
- Salem, K.G.; Tantawy, M.A.; Gawish, A.A.; Gomaa, S.; El-hoshoudy, A.N. Nanoparticles assisted polymer flooding: Comprehensive assessment and empirical correlation. Geoenergy Sci. Eng. 2023, 226, 211753. [Google Scholar] [CrossRef]
- Standnes, D.C.; Skjevrak, I. Literature review of implemented polymer field projects. J. Pet. Sci. Eng. 2014, 122, 761–775. [Google Scholar] [CrossRef]
- She, Y.-H.; Zhang, F.; Xia, J.-J.; Kong, S.-Q.; Wang, Z.-L.; Shu, F.-C.; Hu, J.-M. Investigation of biosurfactant-producing indigenous microorganisms that enhance residue oil recovery in an oil reservoir after polymer flooding. Appl. Biochem. Biotechnol. 2011, 163, 223–234. [Google Scholar] [CrossRef]
- Hou, J.; Song, K.; Wen, Y. Development Direction of Enhanced Oil Recovery Techniques in Polymer Flooded Mature Oilfields. Futur. Technol. 2023, 2, 47–61. [Google Scholar]
- Ren, G.; Wang, J.; Qu, L.; Li, W.; Hu, M.; Bian, L.; Zhang, Y.; Le, J.; Dou, X.; Chen, X.; et al. Compositionsand co-occurrence patterns of bacterial communities associated with polymer-and ASP-flooded petroleum reservoir blocks. Front. Microbiol. 2020, 11, 580363. [Google Scholar] [CrossRef]
- Cui, K.; Sun, S.; Xiao, M.; Liu, T.; Xu, Q.; Dong, H.; Wang, D.; Gong, Y.; Sha, T.; Hou, J.; et al. Microbial mineralization of montmorillonite in low-permeability oil reservoirs for microbial enhanced oil recovery. Appl. Environ. Microbiol. 2018, 84, e00176-18. [Google Scholar] [CrossRef] [PubMed]
- Geetha, S.J.; Banat, I.M.; Joshi, S.J. Biosurfactants: Production and potential applications in microbial enhanced oil recovery (MEOR). Biocatal. Agric. Biotechnol. 2018, 14, 23–32. [Google Scholar]
- She, H.; Kong, D.; Li, Y.; Hu, Z.; Guo, H. Recent advance of microbial enhanced oil recovery (MEOR) in China. Geofuids 2019, 2019, 1871392. [Google Scholar] [CrossRef]
- Gong, X.C.; Liu, Z.S.; Guo, P.; Chi, C.Q.; Chen, J.; Wang, X.B.; Tang, Y.Q.; Wu, X.L.; Liu, C.Z. Bacteria in crude oil survived autoclaving and stimulated differentially by exogenous bacteria. PLoS ONE 2012, 7, e40842. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Xiu, J.; Yu, L.; Huang, L.; Yi, L.; Ma, Y. Biosurfactant production by Bacillus subtilis SL and its potential for enhanced oil recovery in low permeability reservoirs. Sci. Rep. 2022, 12, 7785. [Google Scholar] [CrossRef] [PubMed]
- Markande, A.R.; Patel, D.; Varjani, S. A review on biosurfactants: Properties, applications and current developments. Bioresour. Technol. 2021, 330, 124963. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Du, Z.; Cui, Q.; Dong, H.; Wang, F.; He, P.; Tang, Y. Biosurfactant produced by novel Pseudomonas sp. WJ6 with biodegradation of n-alkanes and polycyclic aromatic hydrocarbons. J. Hazard. Mater. 2014, 276, 489–498. [Google Scholar] [CrossRef]
- Suri, N.; Gassara, F.; Stanislav, P.; Voordouw, G. Microbially enhanced oil recovery by alkylbenzene-oxidizing nitrate-reducing bacteria. Front. Microbiol. 2019, 10, 453830. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-F.; Chen, J.; Liu, Z.-L.; Shou, L.-B.; Lin, D.-D.; Zhou, L.; Yang, S.-Z.; Liu, J.-F.; Li, W.; Gu, J.-D.; et al. Anaerobic degradation of paraffins by thermophilic Actinobacteria under methanogenic conditions. Environ. Sci. Technol. 2020, 54, 10610–10620. [Google Scholar] [CrossRef]
- Vigneron, A.; Alsop, E.B.; Lomans, B.P.; Kyrpides, N.C.; Head, I.M.; Tsesmetzis, N. Succession in the petroleum reservoir microbiome through an oil field production lifecycle. ISME J. 2017, 11, 2141–2154. [Google Scholar] [CrossRef]
- Lin, J.H.; Zhang, K.C.; Tao, W.Y.; Wang, D.; Li, S. Geobacillus strains that have potential value in microbial enhanced oil recovery. Appl. Microbiol. Biotechnol. 2019, 103, 8339–8350. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Li, G.; Le, J.; Liu, X.; Liu, F.; Ma, T. Succession of microbial communities and changes of incremental oil in a post-polymer flooded reservoir with nutrient stimulation. Appl. Microbiol. Biotechnol. 2018, 102, 2007–2017. [Google Scholar] [CrossRef] [PubMed]
- Harvey, R.W.; Harms, H.; Landkamer, L. Transport of microorganisms in the terrestrial subsurface: In situ and laboratory methods. Man. Environ. Microbiol. 2007, 70, 872–897. [Google Scholar]
- Sheehy, A. Oil Recovery Using Microorganisms. WO1992001780A1, 6 February 1992. [Google Scholar]
- Gao, P.; Tian, H.; Li, G.; Zhao, F.; Xia, W.; Gu, J.D.; Le, J.; Ma, T. Environmental conditions and diffusion-limited microbial transfer drive specific microbial communities detected at different sections in oil-production reservoir with water-flooding. bioRxiv 2020, 2020-12. [Google Scholar]
- Sun, G.Z.; Qian, Q.; Hu, J.; Wang, Z.L.; Li, X.M.; Wang, W.D. Study on the Oil Recovery Mechanism of Endogenous Microorganisms in the Z3 Oil Reservoir under Different Types of Activation Agents. J. Xi’an Shiyou Univ. Nat. Sci. Ed. 2019, 2, 8. [Google Scholar]
- Yuan, S.Y.; Wang, Q. New progress and prospect of oilfields development technologies in China. Petrol. Explor. Dev. 2018, 4, 698–711. [Google Scholar] [CrossRef]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef]
- Gloor, G.B.; Hummelen, R.; Macklaim, J.M.; Dickson, R.J.; Fernandes, A.D.; MacPhee, R.; Reid, G. Microbiome profiling by illumina sequencing of combinatorial sequence-tagged PCR products. PLoS ONE 2010, 5, e15406. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Sansupa, C.; Wahdan, S.F.M.; Hossen, S.; Disayathanoowat, T.; Wubet, T.; Purahong, W. Can we use functional annotation of prokaryotic taxa (FAPROTAX) to assign the ecological functions of soil bacteria? Appl. Sci. 2021, 11, 688. [Google Scholar] [CrossRef]
- Zhao, F.; Li, P.; Guo, C.; Shi, R.J.; Zhang, Y. Bioaugmentation of oil reservoir indigenous Pseudomonas aeruginosa to enhance oil recovery through in-situ biosurfactant production without air injection. Bioresour. Technol. 2018, 251, 295–302. [Google Scholar] [CrossRef]
- Gao, W.; Gao, X.; Mi, T.; Han, B.; Zhang, Y.; Li, X.; Yin, X.; Sun, C.; Li, Q.; Cui, Z.; et al. Degradation potential and diversity of oil-degrading bacteria isolated from the sediments of the Jiaozhou Bay, China. Acta Oceanol. Sin. 2019, 38, 54–64. [Google Scholar] [CrossRef]
- Seright, R.S.; Wang, D. Polymer flooding: Current status and future directions. Pet. Sci. 2023, 20, 910–921. [Google Scholar] [CrossRef]
- Shi, M.; Wei, L.; Liao, G.; Han, P.; Hou, Z.; Chen, X.; Wang, Y. Laboratory study on MEOR after polymer flooding. In SPE International Improved Oil Recovery Conference in Asia Pacific; SPE: Kuala Lumpur, Malaysia, 2003; p. SPE-84865. [Google Scholar]
- Patel, J.; Borgohain, S.; Kumar, M.; Rangarajan, V.; Somasundaran, P.; Sen, R. Recent developments in microbial enhanced oil recovery. Renew. Sustain. Energy Rev. 2015, 52, 1539–1558. [Google Scholar] [CrossRef]
- Mokhtari, M.; Ozbayoglu, E.M. Laboratory investigation on gelation behavior of xanthan crosslinked with borate intended to combat lost circulation. In SPE International Production and Operations Conference and Exhibition; SPE: Tunis, Tunisia, 2010; p. SPE-136094. [Google Scholar]
- Sun, G.Z.; Song, X.; Wu, X.L.; Wang, W.D.; Li, X.M.; Wang, Z.L. Study on Activation of Endogenous Functional Microorganisms in Block 3 of Oil-Contaminated Areas by Different Components of Long-Acting Activators. J. Xi’an Shiyou Univ. (Nat. Sci. Ed.) 2017, 32, 112–118. [Google Scholar]
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, F.; Li, H.; Yu, H. Petroleum hydrocarbon-degrading bacteria for the remediation of oil pollution under aerobic conditions: A perspective analysis. Front. Microbiol. 2018, 9, 2885. [Google Scholar] [CrossRef] [PubMed]
- Yahai, Z.J.L. Research progress on interspecies electron transfer of methane-producing microorganisms through mutual oxidation. Microbiol. Bull. 2015, 42, 920–927. [Google Scholar] [CrossRef]
- Youssef, N.; Elshahed, M.S.; McInerney, M.J. Microbial processes in oil fields: Culprits, problems, and opportunities. Adv. Appl. Microbiol. 2009, 66, 141–251. [Google Scholar] [PubMed]
- Varjani, S.J.; Upasani, V.N. Core flood study for enhanced oil recovery through ex-situ bioaugmentation with thermo-and halo-tolerant rhamnolipid produced by Pseudomonas aeruginosa NCIM 5514. Bioresour. Technol. 2016, 220, 175–182. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef]
- Ren, H.; Xiong, S.; Gao, G.; Song, Y.; Cao, G.; Zhao, L.; Zhang, X. Bacteria in the injection water differently impacts the bacterial communities of production wells in high-temperature petroleum reservoirs. Front. Microbiol. 2015, 6, 505. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Hao, B.; Cao, G.; Wang, J.; Feng, Y.; Tan, X.; Wang, W. A study on the microbial community structure in oil reservoirs developed by water flooding. J. Pet. Sci. Eng. 2014, 122, 354–359. [Google Scholar] [CrossRef]
- Beckmann, S.; Lueders, T.; Krüger, M.; von Netzer, F.; Engelen, B.; Cypionka, H. Acetogens and acetoclastic Methanosarcinales govern methane formation in abandoned coal mines. Appl. Environ. Microbiol. 2011, 77, 3749–3756. [Google Scholar] [CrossRef] [PubMed]
- Lambo, A.J.; Patel, T.R. Cometabolic degradation of polychlorinated biphenyls at low temperature by psychrotolerant bacterium Hydrogenophaga sp. IA3-A. Curr. Microbiol. 2006, 53, 48–52. [Google Scholar] [CrossRef]
- Joshi, S.J.; Abed, R.M. Biodegradation of polyacrylamide and its derivatives. Environ. Process. 2017, 4, 463–476. [Google Scholar] [CrossRef]


| Title 1 | K2HPO4 g/L | KH2PO4 g/L | MgSO4 g/L | NH4Cl g/L | CaCl2 g/L | FeCl3 |
|---|---|---|---|---|---|---|
| Scheme 1 | 1.00 | 1.00 | 0.25 | 0.75 | 0.03 | trace |
| Scheme 2 | 1.00 | 1.00 | 0.50 | 0.75 | 0.01 | trace |
| Scheme 3 | 1.00 | 1.00 | 0.75 | 0.75 | 0.02 | trace |
| Scheme 4 | 1.00 | 1.00 | 0.25 | 1.50 | 0.02 | trace |
| Scheme 5 | 1.00 | 1.00 | 0.50 | 1.50 | 0.03 | trace |
| Scheme 6 | 1.00 | 1.00 | 0.75 | 1.50 | 0.01 | trace |
| Scheme 7 | 1.00 | 1.00 | 0.25 | 2.25 | 0.01 | trace |
| Scheme 8 | 1.00 | 1.00 | 0.50 | 2.25 | 0.02 | trace |
| Scheme 9 | 1.00 | 1.00 | 0.75 | 2.25 | 0.03 | trace |
| Index | Medium | Increase (%) | |
|---|---|---|---|
| rev_WJY_JO Medium | rev_FA1_JO Medium | ||
| degradation rate (%) | 14.38 | 20.95 | 45.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Wang, M.; Wei, H.; Wu, X.; Hou, Z.; Zhang, X.; Yang, E. Research on the Functional Microbe Activation System in a Post-Polymer Flooded Reservoir. Processes 2024, 12, 967. https://doi.org/10.3390/pr12050967
Liu Y, Wang M, Wei H, Wu X, Hou Z, Zhang X, Yang E. Research on the Functional Microbe Activation System in a Post-Polymer Flooded Reservoir. Processes. 2024; 12(5):967. https://doi.org/10.3390/pr12050967
Chicago/Turabian StyleLiu, Yinsong, Min Wang, Haiwen Wei, Xiaolin Wu, Zhaowei Hou, Xiumei Zhang, and Erlong Yang. 2024. "Research on the Functional Microbe Activation System in a Post-Polymer Flooded Reservoir" Processes 12, no. 5: 967. https://doi.org/10.3390/pr12050967




