Rapid Acidification and Off-Flavor Reduction of Pea Protein by Fermentation with Lactic Acid Bacteria and Yeasts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pea Protein
2.2. Materials and Chemicals
2.3. Microbial Strains
2.4. Cultivation of Microbial Strains and Inoculum Preparation
2.5. Preparation of Fermentation Mixture
2.6. Development of a Rapid Acidification Test of Pea Protein
2.7. Batch Fermentation
2.8. Biochemical Analysis
2.9. Instrumental Aroma Analysis by HS-SPME-GC-MS
2.10. Sensory Analysis
2.11. Statistical Analysis
3. Results
3.1. Qualitative Assessment of the Acidification Capability of Homofermentative LAB in Pea Protein
3.2. Rapid Acidification of Pea Protein by L. lactis LTH 7123
3.3. Fermentation of Pea Protein with Mixed Cultures
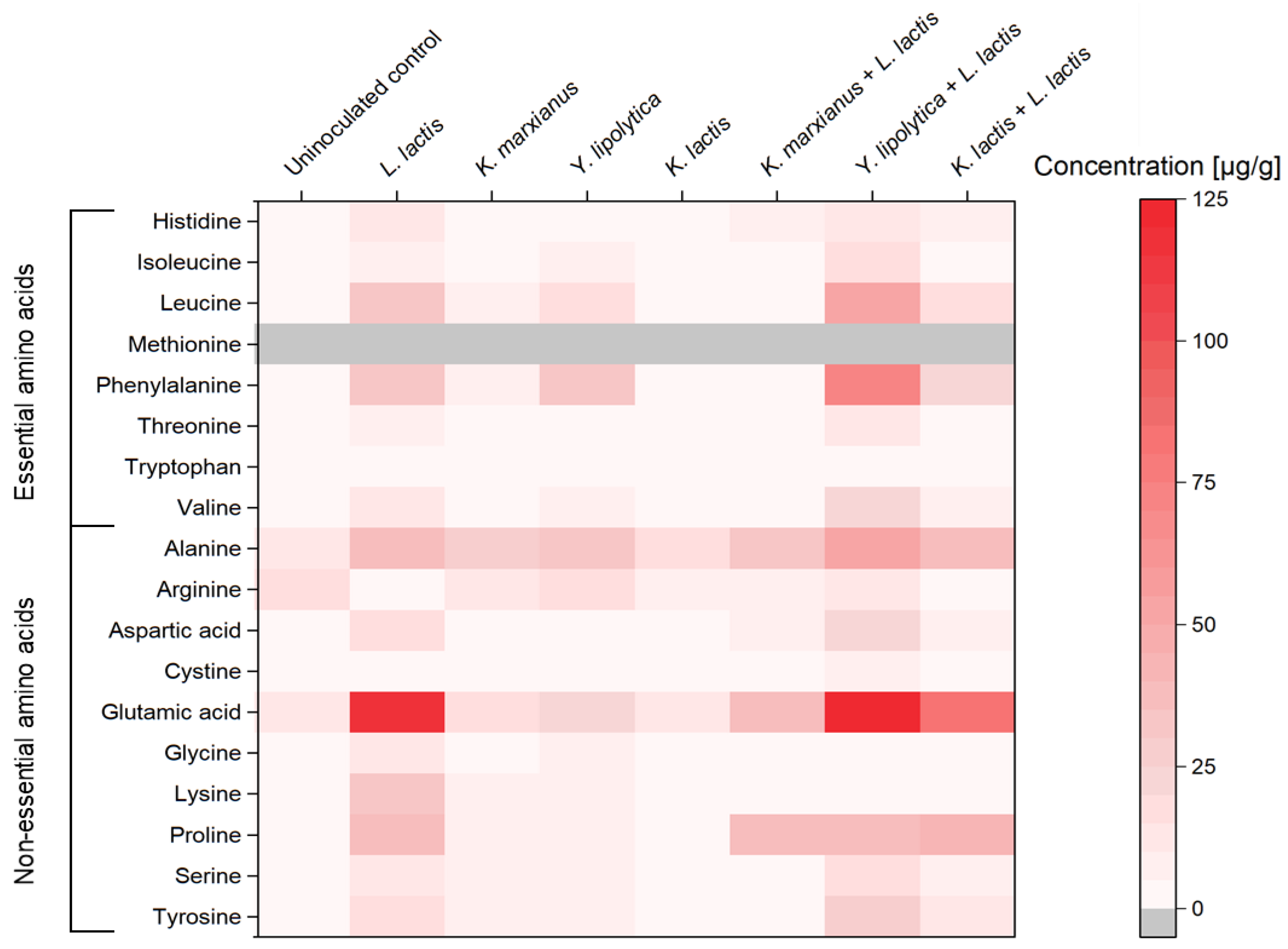
3.4. Free Amino Acids
3.5. Aroma Profile and Sensory Characterization of Fermentation Products
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Delgado, C.L. Rising consumption of meat and milk in developing countries has created a new food revolution. J. Nutr. 2003, 133, 3907S–3910S. [Google Scholar] [CrossRef]
- Aiking, H. Future protein supply. Trends Food Sci. Technol. 2011, 22, 112–120. [Google Scholar] [CrossRef]
- Willett, W.; Rockström, J.; Loken, B.; Springmann, M.; Lang, T.; Vermeulen, S.; Garnett, T.; Tilman, D.; DeClerck, F.; Wood, A.; et al. Food in the anthropocene: The EAT-Lancet Commission on healthy diets from sustainable food systems. Lancet 2019, 393, 447–492. [Google Scholar] [CrossRef]
- Graça, J.; Godinho, C.A.; Truninger, M. Reducing meat consumption and following plant-based diets: Current evidence and future directions to inform integrated transitions. Trends Food Sci. Technol. 2019, 91, 380–390. [Google Scholar] [CrossRef]
- Boye, J.; Zare, F.; Pletch, A. Pulse proteins: Processing, characterization, functional properties and applications in food and feed. Food Res. Int. 2010, 43, 414–431. [Google Scholar] [CrossRef]
- Friedman, M.; Brandon, D.L. Nutritional and health benefits of soy proteins. J. Agric. Food Chem. 2001, 49, 1069–1086. [Google Scholar] [CrossRef] [PubMed]
- García Arteaga, V.; Leffler, S.; Muranyi, I.; Eisner, P.; Schweiggert-Weisz, U. Sensory profile, functional properties and molecular weight distribution of fermented pea protein isolate. Curr. Res. Food Sci. 2021, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Powers, S.E.; Thavarajah, D. Checking agriculture’s pulse: Field pea (Pisum sativum L.), sustainability, and phosphorus use efficiency. Front. Plant Sci. 2019, 10, 1489. [Google Scholar] [CrossRef] [PubMed]
- Lam, A.C.Y.; Can Karaca, A.; Tyler, R.T.; Nickerson, M.T. Pea protein isolates: Structure, extraction, and functionality. Int. Food Res. J. 2018, 34, 126–147. [Google Scholar] [CrossRef]
- Shanthakumar, P.; Klepacka, J.; Bains, A.; Chawla, P.; Dhull, S.B.; Najda, A. The current situation of pea protein and its application in the food industry. Molecules 2022, 27, 5354. [Google Scholar] [CrossRef] [PubMed]
- Kumari, T.; Deka, S.C. Potential health benefits of garden pea seeds and pods: A review. Legum. Sci 2021, 3, e82. [Google Scholar] [CrossRef]
- Millar, K.A.; Gallagher, E.; Burke, R.; McCarthy, S.; Barry-Ryan, C. Proximate composition and anti-nutritional factors of fava-bean (Vicia faba), green-pea and yellow-pea (Pisum sativum) flour. J. Food Compost. Anal. 2019, 82, 103233. [Google Scholar] [CrossRef]
- Gorissen, S.H.M.; Crombag, J.J.R.; Senden, J.M.G.; Waterval, W.A.H.; Bierau, J.; Verdijk, L.B.; van Loon, L.J.C. Protein content and amino acid composition of commercially available plant-based protein isolates. Amino Acids 2018, 50, 1685–1695. [Google Scholar] [CrossRef] [PubMed]
- Dahl, W.J.; Foster, L.M.; Tyler, R.T. Review of the health benefits of peas (Pisum sativum L.). Br. J. Nutr. 2012, 108 (Suppl. S1), S3–S10. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.Y.J.; Srv, A.; Chiang, J.H.; Henry, C.J. Plant proteins for future foods: A roadmap. Foods 2021, 10, 1967. [Google Scholar] [CrossRef] [PubMed]
- Akharume, F.U.; Aluko, R.E.; Adedeji, A.A. Modification of plant proteins for improved functionality: A review. Compr. Rev. Food Sci. Food Saf. 2021, 20, 198–224. [Google Scholar] [CrossRef] [PubMed]
- Fredrikson, M.; Biot, P.; Alminger, M.L.; Carlsson, N.G.; Sandberg, A.S. Production process for high-quality pea-protein isolate with low content of oligosaccharides and phytate. J. Agric. Food Chem. 2001, 49, 1208–1212. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.H.J.; Domoney, C. Perspectives on the genetic improvement of health- and nutrition-related traits in pea. Plant Physiol. Biochem. 2021, 158, 353–362. [Google Scholar] [CrossRef]
- Campos-Vega, R.; Loarca-Piña, G.; Oomah, B.D. Minor components of pulses and their potential impact on human health. Food Res. Int. 2010, 43, 461–482. [Google Scholar] [CrossRef]
- Murray, K.E.; Shipton, J.; Whitfield, F.B.; Last, J.H. The volatiles of off-flavoured unblanched green peas (Pisum sativum). J. Sci. Food Agric. 1976, 27, 1093–1107. [Google Scholar] [CrossRef]
- Trindler, C.; Annika Kopf-Bolanz, K.; Denkel, C. Aroma of peas, its constituents and reduction strategies—Effects from breeding to processing. Food Chem. 2021, 376, 131892. [Google Scholar] [CrossRef]
- Fischer, E.; Cayot, N.; Cachon, R. Potential of microorganisms to decrease the “beany” off-flavor: A review. J. Agric. Food Chem. 2022, 70, 4493–4508. [Google Scholar] [CrossRef]
- Zhang, C.; Hua, Y.; Li, X.; Kong, X.; Chen, Y. Key volatile off-flavor compounds in peas (Pisum sativum L.) and their relations with the endogenous precursors and enzymes using soybean (Glycine max) as a reference. Food Chem. 2020, 333, 127469. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, H.B.; Hansen, M.; Christensen, M.R.; Brockhoff, P.B.; Olsen, C.E. Aroma volatiles of blanched green peas (Pisums ativum L.). J. Agric. Food Chem. 1998, 46, 3727–3734. [Google Scholar] [CrossRef]
- Shen, Y.; Hong, S.; Li, Y. Pea protein composition, functionality, modification, and food applications: A review. Adv. Food Nutr. Res. 2022, 101, 71–127. [Google Scholar] [CrossRef] [PubMed]
- Schindler, S.; Zelena, K.; Krings, U.; Bez, J.; Eisner, P.; Berger, R.G. Improvement of the aroma of pea (Pisum sativum) protein extracts by lactic acid fermentation. Food Biotechnol. 2012, 26, 58–74. [Google Scholar] [CrossRef]
- Engels, W.; Siu, J.; van Schalkwijk, S.; Wesselink, W.; Jacobs, S.; Bachmann, H. Metabolic conversions by lactic acid bacteria during plant protein fermentations. Foods 2022, 11, 1005. [Google Scholar] [CrossRef] [PubMed]
- El Youssef, C.; Bonnarme, P.; Fraud, S.; Péron, A.-C.; Helinck, S.; Landaud, S. Sensory improvement of a pea protein-based product using microbial co-cultures of lactic acid bacteria and yeasts. Foods 2020, 9, 349. [Google Scholar] [CrossRef]
- Ben-Harb, S.; Saint-Eve, A.; Panouillé, M.; Souchon, I.; Bonnarme, P.; Dugat-Bony, E.; Irlinger, F. Design of microbial consortia for the fermentation of pea-protein-enriched emulsions. Int. J. Food Microbiol. 2019, 293, 124–136. [Google Scholar] [CrossRef]
- Emkani, M.; Oliete, B.; Saurel, R. Effect of lactic acid fermentation on legume protein properties, a review. Fermentation 2022, 8, 244. [Google Scholar] [CrossRef]
- Klost, M.; Drusch, S. Structure formation and rheological properties of pea protein-based gels. Food Hydrocoll. 2019, 94, 622–630. [Google Scholar] [CrossRef]
- Lu, Z.X.; He, J.F.; Zhang, Y.C.; Bing, D.J. Composition, physicochemical properties of pea protein and its application in functional foods. Crit. Rev. Food Sci. Nutr. 2020, 60, 2593–2605. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Commission regulation (EC) No 152/2009 of laying down the methods of sampling and analysis for the official control of feed. OJEU 2009, 54, 23–32. [Google Scholar]
- Burdock, G.A. Fenaroli’s Handbook of Flavor Ingredients; CRC Press: Boca Raton, FL, USA, 2016; ISBN 9780429150838. [Google Scholar]
- Rigling, M.; Heger, F.; Graule, M.; Liu, Z.; Zhang, C.; Ni, L.; Zhang, Y. A robust fermentation process for natural chocolate-like flavor production with Mycetinis scorodonius. Molecules 2022, 27, 2503. [Google Scholar] [CrossRef]
- Amoore, J.E.; Hautala, E. Odor as an aid to chemical safety: Odor thresholds compared with threshold limit values and volatilities for 214 industrial chemicals in air and water dilution. J. Appl. Toxicol. 1983, 3, 272–290. [Google Scholar] [CrossRef]
- Chen, J.; Yang, Y.; Deng, Y.; Liu, Z.; Xie, J.; Shen, S.; Yuan, H.; Jiang, Y. Aroma quality evaluation of Dianhong black tea infusions by the combination of rapid gas phase electronic nose and multivariate statistical analysis. LWT 2022, 153, 112496. [Google Scholar] [CrossRef]
- Ruth, J.H. Odor thresholds and irritation levels of several chemical substances: A review. Am. Ind. Hyg. Assoc. J. 1986, 47, A-142–A-151. [Google Scholar] [CrossRef]
- Ebeler, S.E. Gas chromatographic analysis of wine. In Gas Chromatography; Elsevier: Amsterdam, The Netherlands, 2021; pp. 807–833. ISBN 9780128206751. [Google Scholar]
- Nedele, A.-K.; Gross, S.; Rigling, M.; Zhang, Y. Reduction of green off-flavor compounds: Comparison of key odorants during fermentation of soy drink with Lycoperdon pyriforme. Food Chem. 2021, 334, 127591. [Google Scholar] [CrossRef]
- Leffingwell & Associates. Odor & Flavor Detection Thresholds in Water (in Parts per Billion). Available online: https://www.leffingwell.com/odorthre.htm (accessed on 1 December 2023).
- Rigling, M.; Yadav, M.; Yagishita, M.; Nedele, A.-K.; Sun, J.; Zhang, Y. Biosynthesis of pleasant aroma by enokitake (Flammulina velutipes) with a potential use in a novel tea drink. LWT 2021, 140, 110646. [Google Scholar] [CrossRef]
- Liang, J.; Stöppelmann, F.; Schoenbach, J.; Rigling, M.; Nedele, A.-K.; Zhang, Y.; Hannemann, L.; Hua, N.; Heimbach, J.; Kohlus, R.; et al. Influence of peeling on volatile and non-volatile compounds responsible for aroma, sensory, and nutrition in ginger (Zingiber officinale). Food Chem. 2023, 419, 136036. [Google Scholar] [CrossRef]
- Liang, J.; Shalaby, N.; Rigling, M.; Wagner, T.; Heimbach, J.; Fries, A.; Kohlus, R.; Zhang, Y. Characterization of hay-like off-odor in basil samples after various processing and strategies for reducing the off-odor. Food Res. Int. 2022, 162, 112080. [Google Scholar] [CrossRef]
- Sabaté, J.; Soret, S. Sustainability of plant-based diets: Back to the future. Am. J. Clin. Nutr. 2014, 100 (Suppl. 1), 476S–482S. [Google Scholar] [CrossRef]
- Tangyu, M.; Muller, J.; Bolten, C.J.; Wittmann, C. Fermentation of plant-based milk alternatives for improved flavour and nutritional value. Appl. Microbiol. Biotechnol. 2019, 103, 9263–9275. [Google Scholar] [CrossRef]
- Fu, X.; Li, P.; Zhang, L.; Li, S. Understanding the stress responses of Kluyveromyces marxianus after an arrest during high-temperature ethanol fermentation based on integration of RNA-Seq and metabolite data. Appl. Microbiol. Biotechnol. 2019, 103, 2715–2729. [Google Scholar] [CrossRef]
- Dallagnol, A.M.; Pescuma, M.; de Valdez, G.F.; Rollán, G. Fermentation of quinoa and wheat slurries by Lactobacillus plantarum CRL 778: Proteolytic activity. Appl. Microbiol. Biotechnol. 2013, 97, 3129–3140. [Google Scholar] [CrossRef]
- Kurihara, K. Umami the fifth basic taste: History of studies on receptor mechanisms and role as a food flavor. Biomed. Res. Int. 2015, 2015, 189402. [Google Scholar] [CrossRef] [PubMed]
- Forler, B.; Horstmann, G.; Schäfer, J.; Michel, C.; Weiss, A.; Stressler, T.; Fischer, L.; Hinrichs, J.; Schmidt, H. Effects of protein, calcium, and pH on gene transcription, cell-envelope peptidase activity of Lactococcus lactis strains, and the formation of bitter peptides. Foods 2021, 10, 1588. [Google Scholar] [CrossRef] [PubMed]
- Ciurko, D.; Neuvéglise, C.; Szwechłowicz, M.; Lazar, Z.; Janek, T. Comparative analysis of the alkaline proteolytic enzymes of Yarrowia clade species and their putative applications. Int. J. Mol. Sci. 2023, 24, 6514. [Google Scholar] [CrossRef] [PubMed]
- Vong, W.C.; Liu, S.-Q. Changes in volatile profile of soybean residue (okara) upon solid-state fermentation by yeasts. J. Sci. Food Agric. 2017, 97, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Hao, Z.; Wang, R.; Chen, Z.; Zuo, F.; Wan, Y.; Guo, S. Changes of hexanal content in fermented soymilk: Induced by lactic acid bacterial fermentation and thermal treatment. Food Process. Preserv. 2022, 46, e16555. [Google Scholar] [CrossRef]
- Pires, E.J.; Teixeira, J.A.; Brányik, T.; Vicente, A.A. Yeast: The soul of beer’s aroma—A review of flavour-active esters and higher alcohols produced by the brewing yeast. Appl. Microbiol. Biotechnol. 2014, 98, 1937–1949. [Google Scholar] [CrossRef] [PubMed]
- Peddie, H.A.B. Ester formation in brewery fermentations. J. Inst. Brew. 1990, 96, 327–331. [Google Scholar] [CrossRef]
- Zinjarde, S.S. Food-related applications of Yarrowia lipolytica. Food Chem. 2014, 152, 1–10. [Google Scholar] [CrossRef]
- Jach, M.E.; Malm, A. Yarrowia lipolytica as an alternative and valuable source of nutritional and bioactive compounds for humans. Molecules 2022, 27, 2300. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-K.; Ledesma-Amaro, R. What makes Yarrowia lipolytica well suited for industry? Trends Biotechnol. 2023, 41, 242–254. [Google Scholar] [CrossRef]
- Groenewald, M.; Boekhout, T.; Neuvéglise, C.; Gaillardin, C.; van Dijck, P.W.M.; Wyss, M. Yarrowia lipolytica: Safety assessment of an oleaginous yeast with a great industrial potential. Crit. Rev. Microbiol. 2014, 40, 187–206. [Google Scholar] [CrossRef]
- Turck, D.; Castenmiller, J.; de Henauw, S.; Hirsch-Ernst, K.-I.; Kearney, J.; Maciuk, A.; Mangelsdorf, I.; McArdle, H.J.; Naska, A.; Pelaez, C.; et al. Safety of Yarrowia lipolytica yeast biomass as a novel food pursuant to regulation (EU) 2015/2283. EFSA J. 2019, 17, e05594. [Google Scholar] [CrossRef]
- Turck, D.; Bohn, T.; Castenmiller, J.; de Henauw, S.; Hirsch-Ernst, K.I.; Maciuk, A.; Mangelsdorf, I.; McArdle, H.J.; Naska, A.; Pelaez, C.; et al. Safety of an extension of use of Yarrowia lipolytica yeast biomass as a novel food pursuant to regulation (EU) 2015/2283. EFSA J. 2022, 20, e07450. [Google Scholar] [CrossRef]




| Strain | Alternative Strain Designation |
|---|---|
| Bacteria | |
| Latilactobacillus curvatus LTH 955T | DSM 20019, ATCC 25601 |
| Lacticaseibacillus rhamnosus LTH 1409T | DSM 20021, ATCC 7469 |
| Pediococcus acidilactici LTH 1412T | DSM 20284 |
| Latilactobacillus sakei subsp. sakei LTH 3745T | DSM 20017, ATCC 15521 |
| Lactiplantibacillus plantarum LTH 6724 | |
| Pediococcus pentosaceus LTH 6727 | |
| Lactococcus cremoris LTH 7122 | |
| Lactococcus lactis subsp. lactis LTH 7123 | |
| Streptococcus thermophilus LTH 7138 | |
| Lactobacillus delbrueckii subsp. bulgaricus LTH 7139 | |
| Lactococcus lactis subsp. lactis LTH 7163 | DSM 20729, ATCC 11454 |
| Yeasts | |
| Kluyveromyces marxianus LTH 6039 | DSM 70343 |
| Yarrowia lipolytica LTH 6056 | |
| Kluyveromyces lactis LTH 7165 | DSM 70799, ATCC 8585 |
| Compound | Odor Description 1 | Odor Threshold in Water 2 [µg/L] |
|---|---|---|
| Decanal | green | 0.10 |
| Heptanal | floral, green | 3.00 |
| Nonanal | citrus-like | 1.00 |
| (E)-2-Octenal | green | 3.00 |
| 1-Octen-3-ol | mushroom-like | 1.00 |
| 2-Nonanone | grass, fruity, floral [34] | 200.00 |
| 2-Pentylfuran | beany | 6.00 |
| Hexanal | grassy, green | 5.00 |
| (E,E)-2,4-Decadienal | green | 0.07 |
| Ethyl hexanoate | fruity | 1.00 |
| 1-Hexanol | floral, green, perfume-like | 2500.00 |
| Isoamyl acetate | sweetish, banana-like [35] | 17.00 [36] |
| 2-Methylfuran | cocoa-like, nutty [37] | 90.00 [38] |
| 1-Octanol | artificial | 130.00 |
| 2-Isobutyl-3-methoxypyrazine | bell pepper-like, spicy | 0.01 [39] |
| Strains | L-Lactic Acid [g/L] | Ethanol [g/L] |
|---|---|---|
| L. lactis LTH 7123 + K. marxianus LTH 6039 | 2.6 ± 0.1 a | 2.2 ± 0.1 d |
| L. lactis LTH 7123 + Y. lipolytica LTH 6056 | 2.6 ± 0.4 a | 0.3 ± 0.1 |
| L. lactis LTH 7123 + K. lactis LTH 7165 | 2.9 ± 0.2 a | 1.5 ± 0.1 d |
| K. marxianus LTH 6039 | 0.1 ± 0.0 b | 4.3 ± 0.6 c |
| Y. lipolytica LTH 6056 | 0.1 ± 0.0 b | < 0.001 |
| K. lactis LTH 7165 | 0.1 ± 0.0 b | 4.2 ± 1.1 c |
| L. lactis LTH 7123 | 2.4 ± 0.5 a | <0.001 |
| Uninoculated control | 0.1 ± 0.0 b | <0.001 |
| Compound | Uninoculated Control | L. lactis | K. marxianus | Y. lipolytica | K. lactis | K. marxianus + L. lactis | Y. lipolytica + L. lactis | K. lactis + L. lactis |
|---|---|---|---|---|---|---|---|---|
| Decanal | 1.0 ± 0.4 | 2.6 ± 0.2 | 0.3 ± 0.1 | 0.1 ± 0.0 | 0.2 ± 0.0 | 0.3 ± 0.0 | 1.5 ± 0.5 | 0.7 ± 0.2 |
| Heptanal | 110.6 ± 7.0 | 151.0 ± 6.8 | 25.7 ± 7.7 | 0.4 ± 0.1 | 59.7 ± 7.3 | 38.0 ± 3.5 | 1.0 ± 0.2 | 66.0 ± 5.6 |
| Nonanal | 0.6 ± 0.1 | 17.7 ± 1.9 | 0.1 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.2 ± 0.1 | 0.1 ± 0.0 |
| (E)-2-Octenal | 1.0 ± 0.2 | 27.3 ± 2.8 | 1.1 ± 0.6 | 0.1 ± 0.0 | 0.0 ± 0.0 | 1.2 ± 0.0 | 0.2 ± 0.1 | 0.0 ± 0.0 |
| 1-Octen-3-ol | 8.1 ± 0.6 | 38.6 ± 1.9 | 1.0 ± 0.4 | 3.7 ± 0.4 | 0.6 ± 0.1 | 2.0 ± 0.3 | 8.0 ± 1.1 | 2.0 ± 0.3 |
| 2-Nonanone | 3.4 ± 0.2 | 8.4 ± 0.7 | 0.5 ± 0.2 | 0.2 ± 0.1 | 0.3 ± 0.0 | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.5 ± 0.2 |
| 2-Pentylfuran | 283.7 ± 54.8 | 396.3 ± 23.7 | 329.9 ± 44.8 | 157.1 ± 23.4 | 238.6 ± 17.2 | 245.5 ± 14.2 | 237.3 ± 64.4 | 253.8 ± 29.0 |
| Hexanal | 615.9 ± 7.1 | 924.9 ± 13.8 | 5.8 ± 1.0 | 1.9 ± 0.4 | 5.7 ± 0.8 | 17.1 ± 4.4 | 63.8 ± 9.6 | 89.4 ± 12.4 |
| (E,E)-2,4-Decadienal | 0.0 ± 0.0 | 1.9 ± 0.7 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Ethyl hexanoate | 0.4 ± 0.01 | 0.8 ± 0.1 | 47.8 ± 3.8 | 1.2 ± 0.3 | 47.9 ± 3.5 | 41.8 ± 3.3 | 2.6 ± 0.5 | 27.0 ± 2.9 |
| 1-Hexanol | 40.0 ± 0.8 | 1073.1 ± 52.0 | 461.2 ± 40.6 | 142.8 ± 14.9 | 485.9 ± 15.0 | 1058.9 ± 78.3 | 829.1 ± 119.3 | 1161.4 ± 107.4 |
| Isoamyl acetate | 0.0 ± 0.0 | 0.1 ± 0.0 | 158.3 ± 15.7 | 0.0 ± 0.0 | 66.1 ± 1.5 | 316.4 ± 14.2 | 0.2 ± 0.1 | 0.6 ± 0.4 |
| 2-Methylfuran | 3.1 ± 0.8 | 5.5 ± 0.7 | 1.4 ± 0.4 | 4.5 ± 1.2 | 1.9 ± 0.2 | 1.7 ± 0.3 | 5.5 ± 1.1 | 2.0 ± 0.3 |
| 1-Octanol | 1.9 ± 0.3 | 21.3 ± 1.8 | 12.2 ± 1.0 | 0.0 ± 0.0 | 2.1 ± 0.0 | 12.0 ± 2.7 | 0.8 ± 0.3 | 8.3 ± 0.9 |
| 2-Isobutyl-3-methoxypyrazine | 0.4 ± 0.2 | 1.0 ± 0.1 | 0.1 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0 |
| Strain | Descriptors |
|---|---|
| L. lactis LTH 7123 + K. marxianus LTH 6039 | fermented, alcoholic, sourish |
| L. lactis LTH 7123 + Y. lipolytica LTH 6056 | yeasty, slightly cereal-like |
| L. lactis LTH 7123 + K. lactis LTH 7165 | fermented, malty, milky |
| K. marxianus LTH 6039 | fruity, malty, dough-like, sweetish |
| Y. lipolytica LTH 6056 | green, cereal-like, beany |
| K. lactis LTH 7165 | fruity, grape pomace-like, alcoholic |
| L. lactis LTH 7123 | cereal-like, soy-like, beany, weakly sourish |
| Uninoculated control | cereal-like, green, soy drink-like, beany |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zipori, D.; Hollmann, J.; Rigling, M.; Zhang, Y.; Weiss, A.; Schmidt, H. Rapid Acidification and Off-Flavor Reduction of Pea Protein by Fermentation with Lactic Acid Bacteria and Yeasts. Foods 2024, 13, 588. https://doi.org/10.3390/foods13040588
Zipori D, Hollmann J, Rigling M, Zhang Y, Weiss A, Schmidt H. Rapid Acidification and Off-Flavor Reduction of Pea Protein by Fermentation with Lactic Acid Bacteria and Yeasts. Foods. 2024; 13(4):588. https://doi.org/10.3390/foods13040588
Chicago/Turabian StyleZipori, Dor, Jana Hollmann, Marina Rigling, Yanyan Zhang, Agnes Weiss, and Herbert Schmidt. 2024. "Rapid Acidification and Off-Flavor Reduction of Pea Protein by Fermentation with Lactic Acid Bacteria and Yeasts" Foods 13, no. 4: 588. https://doi.org/10.3390/foods13040588






