RNA-Seq Reveals That Multiple Pathways Are Involved in Tuber Expansion in Tiger Nuts (Cyperus esculentus L.)
Abstract
:1. Introduction
2. Results
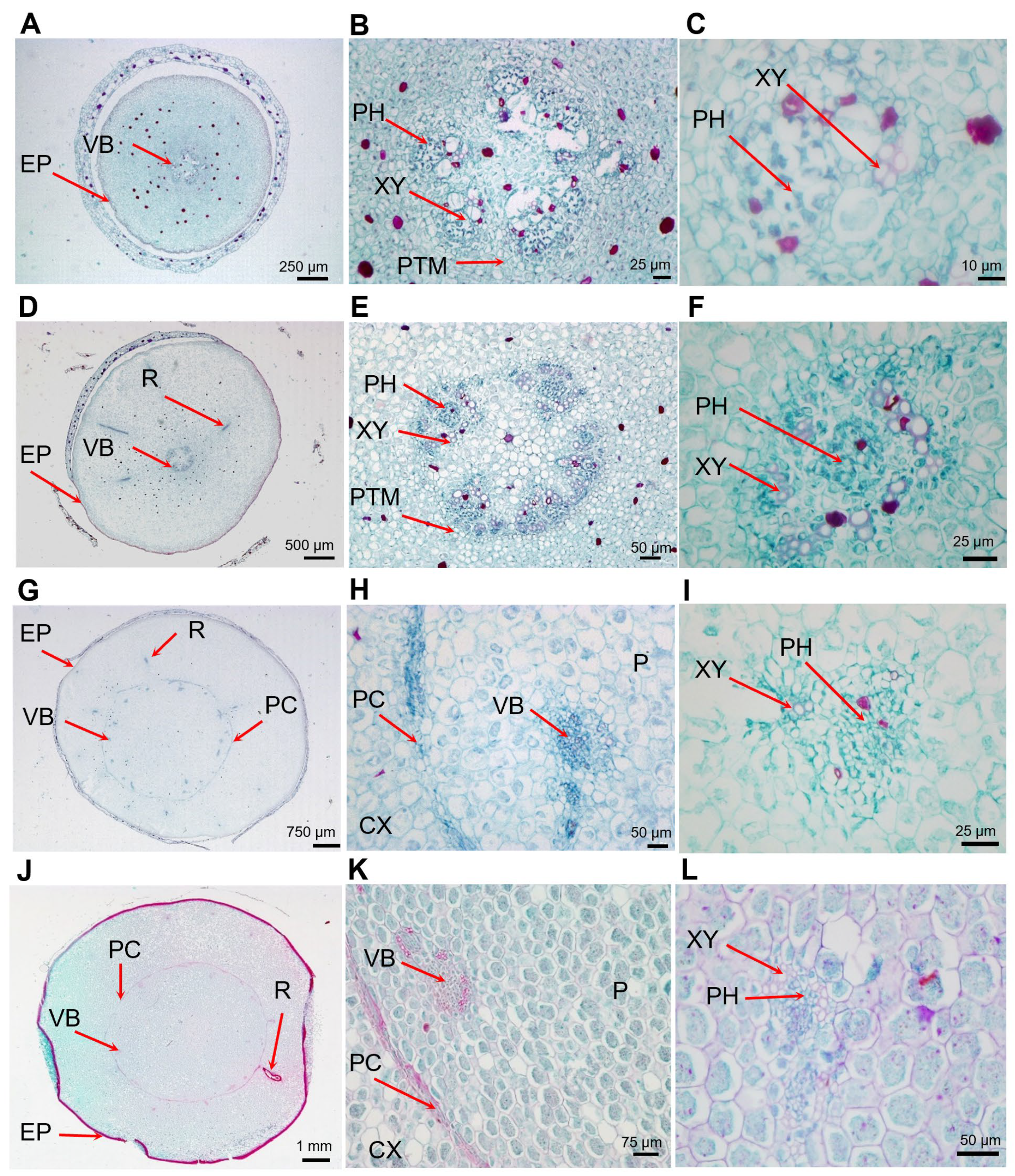
2.1. Characteristics of Tuber Development
2.2. Transcriptome Analysis of Tiger Nut Tubers
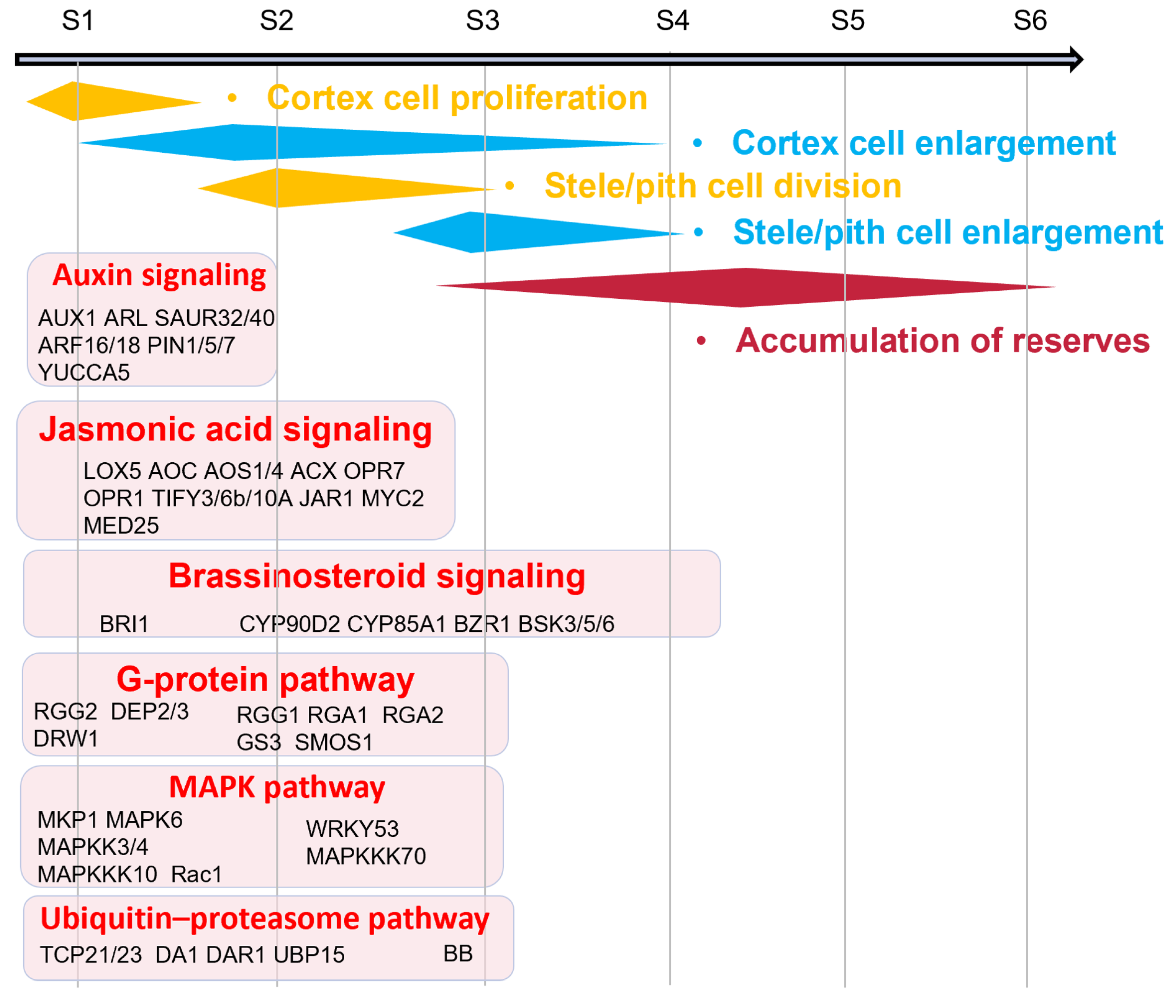
2.3. Cell Division-Related DEGs in Tuber Expansion
2.4. Cell Enlargement-Related DEGs in Tuber Expansion
2.5. Energy-Supply Related DEGs in Tuber Expansion
2.6. Hormone-Related Pathways Regulate Tuber Expansion
2.7. Transcriptional Pathways Regulate Tuber Expansion
2.8. Transcription Factors in Tuber Expansion
2.9. Validation of DEGs Using RT-qPCR
3. Discussion
3.1. Structure of Tiger Nut Tubers
3.2. Characteristics of Tiger Nut Tuber Expansion
3.3. Cell Cycle and Cell Enlargement Controls Tuber Expansion
3.4. The Hormone Signal Transduction Pathway Regulates Tuber Expansion
3.5. Transcriptional Regulatory Pathways Involved in Tuber Expansion
3.6. Characteristics and Commonalities of Tuber Development in Tiger Nut
4. Materials and Methods
4.1. Planting and Sampling
4.2. Preparation of Paraffin Sections
4.3. Determination of Hormone Content
4.4. RNA Extraction, cDNA Library Construction, and RNA-Seq
4.5. Transcriptome Data Analysis
4.6. qRT-PCR Verification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, N.; Li, Y. Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 2016, 33, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Goldy, C.; Barrera, V.; Taylor, I.; Buchensky, C.; Vena, R.; Benfey, P.; De Veylder, L.; Rodriguez, R. SCARECROW-LIKE28 modulates organ growth in Arabidopsis by controlling mitotic cell cycle exit, endoreplication, and cell expansion dynamics. New Phytol. 2023, 237, 1652–1666. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Van Bel, M.; Inze, D.; Baekelandt, A. Leaf growth-complex regulation of a seemingly simple process. Plant J. 2024, 117, 1018–1051. [Google Scholar] [CrossRef]
- Shi, C.; Ren, Y.; Liu, L.; Wang, F.; Zhang, H.; Tian, P.; Pan, T.; Wang, Y.; Jing, R. Ubiquitin specific protease 15 has an important role in regulating grain width and size in rice. Plant Physiol. 2019, 180, 381–391. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiong, Y.; Liu, R.; Xue, H.; Yang, Z. The Rho-family GTPase OsRac1 controls rice grain size and yield by regulating cell division. Proc. Natl. Acad. Sci. USA 2019, 116, 16121–16126. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cai, X.; Zhang, B.; Wu, S.; Wang, R.; Li, N. ERECTA regulates seed size independently of its intracellular domain via MAPK-DA1-UBP15 signaling. Plant Cell 2022, 34, 3773–3789. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gao, F.; Xie, K.; Zeng, X.; Cao, Y.; Zeng, J.; He, Z.; Ren, Y.; Li, W. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotech. J. 2016, 14, 2134–2146. [Google Scholar] [CrossRef]
- Tong, H.; Liu, L.; Jin, Y.; Du, L.; Yin, Y. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-Like kinase to mediate brassinosteroid responses in rice. Plant Cell 2012, 24, 2562–2577. [Google Scholar] [CrossRef]
- Zhu, Y.; Luo, X.; Liu, X.; Wu, W.; Cui, X.; He, Y. Arabidopsis PEAPODs function with LIKE HETEROCHROMATIN PROTEIN1 to regulate lateral organ growth. J. Integr. Plant Biol. 2020, 62, 812–831. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Meng, Y.; Zeng, J.; Luo, Y.; Gao, S. Coordination between GROWTH-REGULATING FACTOR1 and GRF-INTERACTING FACTOR1 plays a key role in regulating leaf growth in rice. BMC Plant Biol. 2020, 20, 200. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, D.; Xue, M. Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J. Integr. Plant Biol. 2014, 56, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lammeren, A.; Vermeer, E.; Vreugdenhil, D. The role of gibberellin, abscisic acid, and sucrose in the regulation of potato tuber formation in vitro. Plant Physiol. 1998, 117, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Begum, S.; Jing, S.; Liu, Y.; Sun, X.; Wang, E.; Kawochar, M.; Qin, J.; Liu, J.; Song, B. Modulation of JA signalling reveals the influence of StJAZ1-like on tuber initiation and tuber bulking in potato. Plant J. 2021, 109, 952–964. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, D.; Ji, H. Sucrose metabolism in developing oil-rich tubers of Cyperus esculentus: Comparative transcriptome analysis. BMC Plant Biol. 2018, 18, 151. [Google Scholar] [CrossRef]
- Bado, S.; Bazongo, P.; Son, G.; Kyaw, M.; Forster, B.; Nielen, S. Physicochemical characteristics and composition of three morphotypes of Cyperus esculentus tubers and tuber oils. J. Anal. Methods Chem. 2015, 2015, 673547. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Liu, D.; Yang, Z. High oil accumulation in tuber of yellow nutsedge compared to purple nutsedge is associated with more abundant expression of genes involved in fatty acid synthesis and triacylglycerol storage. Biotechnol. Biofuels 2021, 14, 54. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Ji, H.; Liu, D. Oil biosynthesis in underground oil-rich storage vegetative tissue: Comparison of Cyperus esculentus tuber with oil seeds and fruits. Plant Cell Physiol. 2016, 57, 2519–2540. [Google Scholar] [CrossRef]
- Zhao, X.; Yi, L.; Ren, Y.; Li, J.; Ren, W.; Hou, Z.; Su, S.; Wang, J.; Zhang, Y.; Dong, Q.; et al. Chromosome-scale genome assembly of the yellow nutsedge (Cyperus esculentus). Genome Biol. Evol. 2023, 15, evad027. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Simmons, C.R. Cell number counts-The fw2.2 and CNR genes and implications for controlling plant fruit and organ size. Plant Sci. 2011, 181, 1–7. [Google Scholar] [CrossRef]
- Ashihara, H.; Stasolla, C.; Fujimura, T.; Crozier, A. Purine salvage in plants. Phytochemistry 2018, 147, 89–124. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Jing, W.; Liang, X.; Wang, Y.; Wang, R.; Zhu, X.; Sun, X. Transcriptome profiling of taproot reveals complex regulatory networks during taproot thickening in Radish (Raphanus sativus L.). Front. Plant Sci. 2016, 7, 1210. [Google Scholar] [CrossRef]
- Sun, X.; Zhuang, Y.; Lin, H.; Zhou, H. Patellin1 negatively regulates plant salt tolerance by attenuating nitric oxide accumulation in Arabidopsis. Plant Sig. Behav. 2019, 14, 1675472. [Google Scholar] [CrossRef]
- Smertenko, A.; Saleh, N.; Igarashi, H.; Mori, H.; Hauser-Hahn, I.; Jiang, C.; Sonobe, S.; Lloyd, C.; Hussey, P.J. A new class of microtubule-associated proteins in plants. Nat. Cell Biol. 2000, 2, 750–753. [Google Scholar] [CrossRef]
- Sajjan, A.; Guggari, A. The effect of seed size on seed recovery and quality in chickpea (Cicer arietinum L.). Legume Res. 2023, 46, 757–761. [Google Scholar] [CrossRef]
- Ku, A.; Huang, Y.; Wang, Y.; Ma, D. IbMADS1 (Ipomoea batatas MADS-box 1 gene) is involved in tuberous root initiation in sweet potato (Ipomoea batatas). Ann. Bot. 2008, 102, 57–67. [Google Scholar] [CrossRef]
- Tang, G.; Lüscher, M.; Sturm, A. Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell 1999, 11, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Guo, Z.; Song, L.; Wang, Y.; Cheng, Y. NCP1/AtMOB1A plays key roles in auxin-mediated Arabidopsis development. PLoS Genet. 2016, 12, e1005923. [Google Scholar] [CrossRef] [PubMed]
- Uji, Y.; Kashihara, K.; Kiyama, H.; Mochizuki, S.; Akimitsu, K.; Gomi, K. Jasmonic acid-induced VQ-Motif-Containing protein OsVQ13 influences the OsWRKY45 signaling pathway and grain size by associating with OsMPK6 in rice. Int. J. Mol. Sci. 2019, 20, 2917. [Google Scholar] [CrossRef] [PubMed]
- Varshney, V.; Hazra, A.; Rao, V.; Ghosh, S.; Kamble, N.; Achary, R. The Arabidopsis F-box protein SKP1-INTERACTING PARTNER 31 modulates seed maturation and seed vigor by targeting JASMONATE ZIM DOMAIN proteins independently of jasmonic acid-isoleucine. Plant Cell 2023, 35, 3712–3738. [Google Scholar] [CrossRef]
- Ko, H.; Hideki, Y.; Koichiro, A.; Mayuko, K.; Makoto, H.; Tokunori, H. SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING form a complex to integrate auxin and brassinosteroid signaling in rice. Mol. Plant. 2017, 10, 590–604. [Google Scholar]
- Yuan, H.; Xu, Z.; Chen, W.; Deng, C.; Liu, Y.; Yuan, M.; Gao, P.; Shi, H.; Tu, B.; Li, T. OsBSK2, a putative brassinosteroid-signalling kinase, positively controls grain size in rice. J. Exp. Bot. 2022, 73, 5529–5542. [Google Scholar] [CrossRef]
- Hu, Y.; Xie, Q.; Chua, N. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 2003, 15, 1951–1961. [Google Scholar] [CrossRef]
- Busov, V.; Brunner, A.; Strauss, S. Genes for control of plant stature and form. New Phytol. 2008, 177, 589–607. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Gerald, J.; Berger, F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 2005, 17, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Huang, H.; Hu, Y.; Zhu, S.; Wang, Z.; Lin, W. Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 2013, 162, 1965–1977. [Google Scholar] [CrossRef]
- Acheampong Atiako, K.; Shanks, C.; Cheng, C. EXO70D isoforms mediate selective autophagic degradation of type-A ARR proteins to regulate cytokinin sensitivity. Proc. Natl. Acad. Sci. USA 2020, 117, 27034–27043. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Tao, Y.; Miao, J.; Wang, J.; Li, W.; Xu, Y. RGG1, involved in the cytokinin regulatory pathway, controls grain size in rice. Rice 2020, 13, 76. [Google Scholar] [CrossRef]
- Utsunomiya, Y.; Samejima, C.; Takayanagi, Y.; Izawa, Y.; Iwasaki, Y. Suppression of the rice heterotrimeric G protein β-subunit gene, RGB1, causes dwarfism and browning of internodes and lamina joint regions. Plant J. 2011, 67, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Chen, K.; Dong, N.; Shi, C.; Ye, W.; Gao, J.; Shan, J.; Lin, H. GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 2018, 30, 871–888. [Google Scholar] [CrossRef]
- Liu, Z.; Mei, E.; Tian, X.; He, M.; Tang, J.; Xu, M.; Liu, J.; Song, L. OsMKKK70 regulates grain size and leaf angle in rice through the OsMKK4-OsMAPK6-OsWRKY53 signaling pathway. J. Integr. Plant Biol. 2021, 63, 2043–2057. [Google Scholar] [CrossRef]
- Ren, D.; Ding, C.; Qian, Q. Molecular bases of rice grain size and quality for optimized productivity. Sci. Bull. 2023, 68, 314–350. [Google Scholar] [CrossRef]
- Hu, L.; Ye, M.; Li, R.; Zhang, T.; Zhou, G.; Wang, Q.; Lu, J.; Lou, Y. The rice transcription factor WRKY53 suppresses herbivore-induced defenses by acting as a negative feedback modulator of mitogen-activated protein kinase activity. Plant Physiol. 2015, 169, 2907–2921. [Google Scholar]
- Bai, X.; Chen, T.; Wu, Y.; Tang, M.; Xu, Z. Selection and validation of reference genes for qRT-PCR analysis in the oil-rich tuber crop tiger nut (Cyperus esculentus) based on transcriptome data. Int. J. Mol. Sci. 2021, 22, 2569. [Google Scholar] [CrossRef]
- Kuznetsova, K.; Dodueva, I.; Pautov, A.; Krylova, E.; Lutova, L. Genetic control of storage root development. Russ. J. Plant Physiol. 2020, 67, 589–605. [Google Scholar] [CrossRef]
- Machaj, G.; Bostan, H.; Macko-Podgórni, A.; Iorizzo, M.; Grzebelus, D. Comparative transcriptomics of root development in wild and cultivated carrots. Genes 2018, 9, 431. [Google Scholar] [CrossRef]
- Villordon, A.; La Bonte, D.; Firon, N.; Kfir, Y.; Pressman, E.; Schwartz, A. Characterization of adventitious root development in sweetpotato. HortScience 2009, 44, 651–655. [Google Scholar] [CrossRef]
- Siebers, T.; Catarino, B.; Agusti, J. Identification and expression analyses of new potential regulators of xylem development and cambium activity in cassava (Manihot esculenta). Planta 2017, 245, 539–548. [Google Scholar] [CrossRef]
- Hearn, D.; O’Brien, P.; Poulsen, T. Comparative transcriptomics reveals shared gene expression changes during independent evolutionary origins of stem and hypocotyl/root tubers in Brassica (Brassicaceae). PLoS ONE 2018, 13, e0197166. [Google Scholar] [CrossRef]
- Artschwager, E. On the anatomy of the sweet potato root, with notes on internal breakdown. J. Agric. Res. 1924, 27, 157–166. [Google Scholar]
- Mehdi, R.; Lamm, C.; Bodampalli, A.; Müdsam, C.; Saeed, M.; Klima, J. Symplasmic phloem unloading and radial post-phloem transport via vascular rays in tuberous roots of Manihot esculenta. J. Exp. Bot. 2019, 70, 5559–5573. [Google Scholar] [CrossRef] [PubMed]
- Zaki, H.; Takahata, Y.; Yokoi, S. Analysis of the morphological and anatomical characteristics of roots in three radish (Raphanus sativus) cultivars that differ in root shape. J. Hortic Sci. Biotec. 2012, 87, 172–178. [Google Scholar] [CrossRef]
- Xu, X.; Vreugdenhil, D.; van Lammeren, A. Cell division and cell enlargement during potato tuber formation. J. Exp. Bot. 1998, 49, 573–582. [Google Scholar] [CrossRef]
- Gifford, E.; Bayer, D. Developmental anatomy of Cyperus esculentus (Yellow nutsedge). Int. J. Mol. Sci. 1995, 156, 622–629. [Google Scholar] [CrossRef]
- Francis, D. The plant cell cycle-15 years on. New Phytol. 2007, 174, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef]
- Roumeliotis, E.; Kloosterman, B.; Oortwijn, M.; Visser, R.; Bachem, C. The PIN family of proteins in potato and their putative role in tuberization. Front. Plant Sci. 2013, 4, 524. [Google Scholar] [CrossRef]
- Borzenkova, R.; Borovkova, P. Developmental patterns of phytohormone content in the cortex and pith of potato tubers as related to their growth and starch content. Russ. J. Plant Physiol. 2003, 50, 119–124. [Google Scholar] [CrossRef]
- Huang, S.; Zheng, C.; Zhao, Y.; Li, Q.; Liu, J.; Deng, R.; Lei, T.; Wang, S.; Wang, X. RNA interference knockdown of the brassinosteroid receptor BRI1 in potato (Solanum tuberosum L.) reveals novel functions for brassinosteroid signaling in controlling tuberization. Sci. Hortic. 2021, 290, 110516. [Google Scholar] [CrossRef]
- Nam, K.; Kong, F.; Matsuura, H.; Takahashi, K.; Nabeta, K.; Yoshihara, T. Temperature regulates tuber-inducing lipoxygenase-derived metabolites in potato (Solanum tuberosum). J. Plant Physiol. 2008, 165, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, J.; Hao, B.; Lu, Y.; Qian, Z.; Li, Y.; Ye, S. Comparative transcriptome and metabolome analyses provide new insights into the molecular mechanisms underlying taproot thickening in Panax notoginseng. BMC Plant Biol. 2019, 19, 451. [Google Scholar] [CrossRef]
- Liu, W.; Xu, X.; Wang, X. Unique morphology of Sarcobatus baileyi male inflorescence and its botanical implications. Plants 2023, 12, 1917. [Google Scholar] [CrossRef] [PubMed]
- Forcat, S.; Bennett, M.; Mansfield, J.; Grant, M. A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods 2008, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Dumenil, J.; Lu, F.; Li, N. Ubiquitylation activates a peptidase that promotes cleavage and destabilization of its activating E3 ligases and diverse growth regulatory proteins to limit cell proliferation in Arabidopsis. Genes Dev. 2017, 31, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Sun, S.; Min, F.; Hu, X.; Zhang, Y.; Cheng, Y.; Li, H.; Wang, X.; Liu, X. Integrating network pharmacology and transcriptomic strategies to explore the pharmacological mechanism of hydroxysafflor yellow A in delaying liver aging. Int. J. Mol. Sci. 2022, 23, 14281. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Zhu, M.; Yu, J.; Han, R.; Tang, C.; Xu, T.; Liu, J.; Li, Z. RNA-Seq and iTRAQ reveal multiple pathways involved in storage root formation and development in sweet potato (Ipomoea batatas L.). BMC Plant Biol. 2019, 19, 136. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.; Livak, K. Analyzing real-time PCR data by the comparative C (T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]







| Stage | Cell Size (μm) | Cortical Thickness (mm) | Stele Radius (mm) | Cortex Thickness:Stele Radius Ratio | |||
|---|---|---|---|---|---|---|---|
| Cortex | Stele | ||||||
| Tangential | Radial | Tangential | Radial | ||||
| S1 | 21.47 ± 2.83 | 15.26 ± 2.19 | 15.21 ± 1.69 | 10.49 ± 1.21 | 0.70 ± 0.01 | 0.17 ± 0.02 | 3.27 |
| S2 | 42.09 ± 6.10 | 28.77 ± 4.62 | 26.89 ± 2.86 | 19.40 ± 2.46 | 1.39 ± 0.01 | 0.23 ± 0.02 | 5.04 |
| S3 | 50.31 ± 5.32 | 36.93 ± 4.83 | 52.74 ± 11.73 | 38.57 ± 8.89 | 1.85 ± 0.01 | 1.97 ± 0.01 | 0.90 |
| S4 | 56.32 ± 9.74 | 39.59 ± 11.31 | 60.47 ± 9.08 | 50.43 ± 11.59 | 2.39 ± 0.02 | 2.70 ± 0.02 | 0.88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, G.; Wu, G.; Jiang, H.; Bai, X.; Chen, Y. RNA-Seq Reveals That Multiple Pathways Are Involved in Tuber Expansion in Tiger Nuts (Cyperus esculentus L.). Int. J. Mol. Sci. 2024, 25, 5100. https://doi.org/10.3390/ijms25105100
Hou G, Wu G, Jiang H, Bai X, Chen Y. RNA-Seq Reveals That Multiple Pathways Are Involved in Tuber Expansion in Tiger Nuts (Cyperus esculentus L.). International Journal of Molecular Sciences. 2024; 25(10):5100. https://doi.org/10.3390/ijms25105100
Chicago/Turabian StyleHou, Guangshan, Guojiang Wu, Huawu Jiang, Xue Bai, and Yaping Chen. 2024. "RNA-Seq Reveals That Multiple Pathways Are Involved in Tuber Expansion in Tiger Nuts (Cyperus esculentus L.)" International Journal of Molecular Sciences 25, no. 10: 5100. https://doi.org/10.3390/ijms25105100





