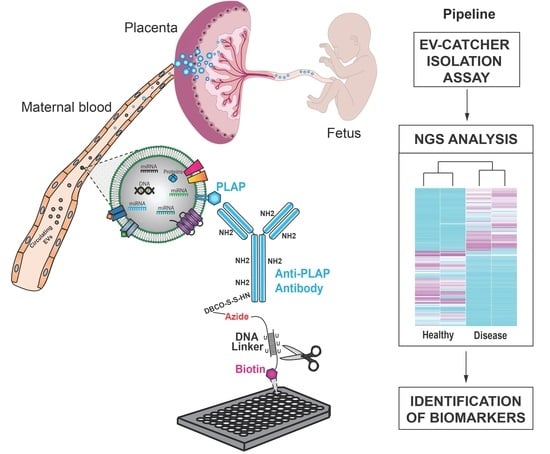
Customizing EV-CATCHER to Purify Placental Extracellular Vesicles from Maternal Plasma to Detect Placental Pathologies
Abstract
:1. Introduction
2. Results
2.1. Validation of PLAP Customization of EV-CATCHER
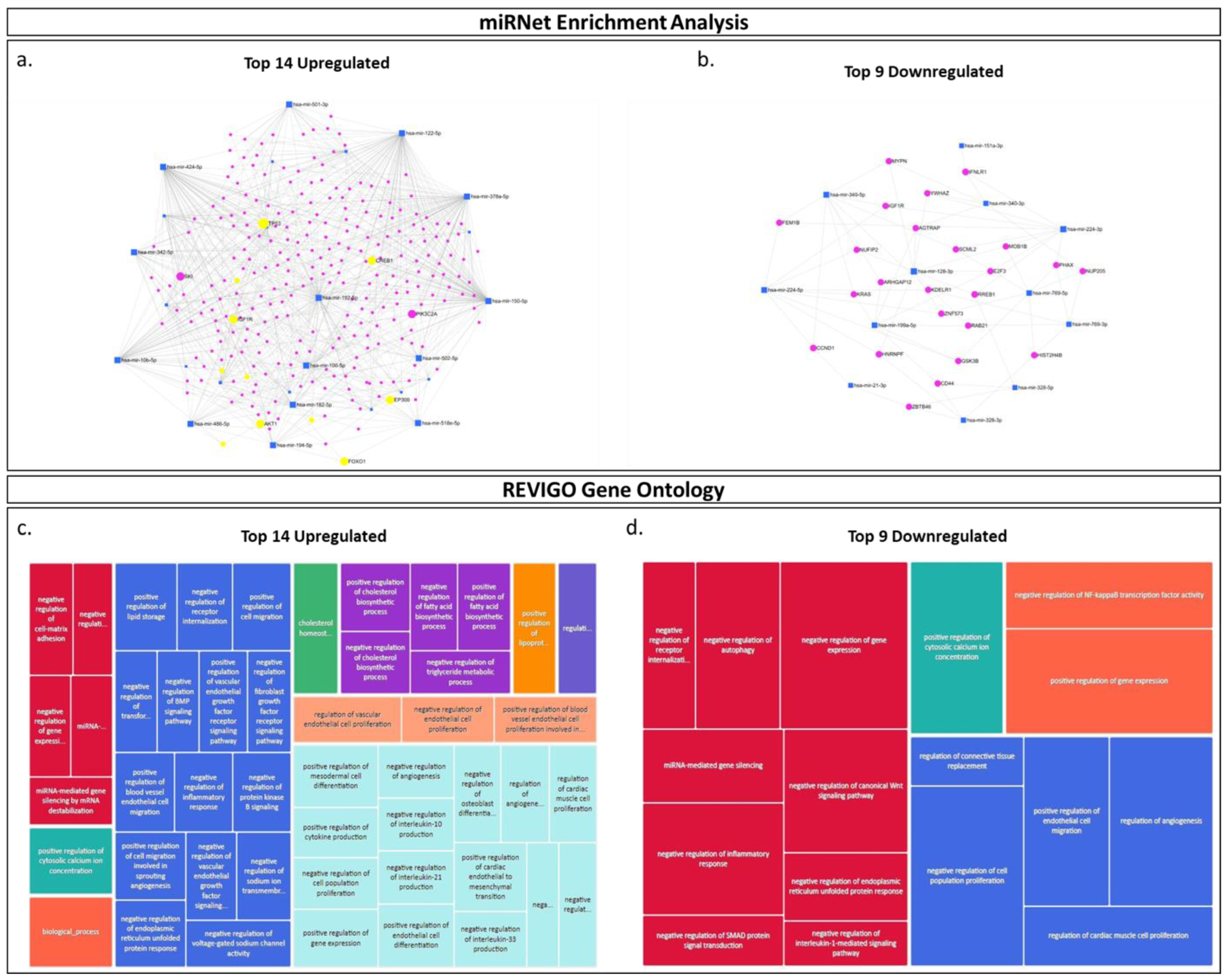
2.2. miRNA Analysis of PLAP+ EVs Purified from Maternal Plasma of Previa and Percreta Pregnancies Using the PLAP Customized EV-CATCHER Assay
2.3. miRNA Pathway Enrichment Analysis and Prediction of Biological Pathway Involvement Using Gene Ontology (GO)
3. Discussion
4. Materials and Methods
4.1. Clinical Specimen Collection
4.2. Western Blot Analysis
4.3. EV-CATCHER Isolation of PLAP+ Small-Extracellular Vesicles
4.4. Transmission Electron Microscopy
4.5. ONi Super Resolution Nanoimaging
4.6. RNA Extractions
4.7. Small-RNA cDNA Library Preparations
4.8. In Silico miRNA Enrichment and Gene Ontology Analyses
4.9. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Afshar, Y.; Dong, J.; Zhao, P.; Li, L.; Wang, S.; Zhang, R.Y.; Zhang, C.; Yin, O.; Han, C.S.; Einerson, B.D.; et al. Circulating trophoblast cell clusters for early detection of placenta accreta spectrum disorders. Nat. Commun. 2021, 12, 4408. [Google Scholar] [CrossRef] [PubMed]
- Morlando, M.; Collins, S. Placenta Accreta Spectrum Disorders: Challenges, Risks, and Management Strategies. Int. J. Women’s Health 2020, 12, 1033–1045. [Google Scholar] [CrossRef] [PubMed]
- Pegu, B.; Thiagaraju, C.; Nayak, D.; Subbaiah, M. Placenta accreta spectrum-a catastrophic situation in obstetrics. Obstet. Gynecol. Sci. 2021, 64, 239–247. [Google Scholar] [CrossRef] [PubMed]
- American College of Obstetricians and Gynecologists; Society for Maternal-Fetal Medicine. Obstetric Care Consensus No. 7: Placenta Accreta Spectrum. Obstet. Gynecol. 2018, 132, e259–e275. [Google Scholar] [CrossRef] [PubMed]
- Bartels, H.C.; Postle, J.D.; Downey, P.; Brennan, D.J. Placenta Accreta Spectrum: A Review of Pathology, Molecular Biology, and Biomarkers. Dis. Markers 2018, 2018, 1507674. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bailit, J.L.; Grobman, W.A.M.; Rice, M.M.; Reddy, U.M.; Wapner, R.J.; Varner, M.W.; Leveno, K.J.; Iams, J.D.; Tita, A.T.; Saade, G.; et al. Morbidly adherent placenta treatments and outcomes. Obstet. Gynecol. 2015, 125, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, K.E.; Sellers, S.; Spark, P.; Kurinczuk, J.J.; Brocklehurst, P.; Knight, M. The management and outcomes of placenta accreta, increta, and percreta in the UK: A population-based descriptive study. BJOG 2014, 121, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Silver, R.M. Abnormal Placentation: Placenta Previa, Vasa Previa, and Placenta Accreta. Obstet. Gynecol. 2015, 126, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Piñas Carrillo, A.; Chandraharan, E. Placenta accreta spectrum: Risk factors, diagnosis and management with special reference to the Triple P procedure. Women’s Health 2019, 15, 1745506519878081. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jauniaux, E.; Ayres-de-Campos, D.; Langhoff-Roos, J.; Fox, K.A.; Collins, S.; Diagnosis, F.P.A. Management Expert Consensus P: FIGO classification for the clinical diagnosis of placenta accreta spectrum disorders. Int. J. Gynaecol. Obstet. 2019, 146, 20–24. [Google Scholar] [CrossRef] [PubMed]
- DaSilva-Arnold, S.C.; Zamudio, S.; Al-Khan, A.; Alvarez-Perez, J.; Mannion, C.; Koenig, C.; Luke, D.; Perez, A.M.; Petroff, M.; Alvarez, M.; et al. Human trophoblast epithelial-mesenchymal transition in abnormally invasive placenta. Biol. Reprod. 2018, 99, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Jauniaux, E.; Bunce, C.; Grønbeck, L.; Langhoff-Roos, J. Prevalence and main outcomes of placenta accreta spectrum: A systematic review and meta-analysis. Am. J. Obstet. Gynecol. 2019, 221, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Di Mascio, D.; Calì, G.; D’antonio, F. Updates on the management of placenta accreta spectrum. Minerva Obstet. Gynecol. 2019, 71, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Kingdom, J.C.; Hobson, S.R.; Murji, A.; Allen, L.; Windrim, R.C.; Lockhart, E.; Collins, S.L.; Majd, H.S.; Alazzam, M.; Naaisa, F.; et al. Minimizing surgical blood loss at Cesarean hysterectomy for placenta previa with evidence of placenta increta or placenta percreta: The state of play in 2020. Am. J. Obstet. Gynecol. 2020, 223, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Jauniaux, E.; Moffett, A.; Burton, G.J. Placental Implantation Disorders. Obstet. Gynecol. Clin. N. Am. 2019, 47, 117–132. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, H.J.; Nippita, T.A.; Torvaldsen, S.; Ibiebele, I.; Ford, J.B.; Patterson, J.A. Outcomes of Subsequent Births after Placenta Accreta Spectrum. Obstet. Gynecol. 2020, 136, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Silver, R.M.; Branch, D.W. Placenta Accreta Spectrum. N. Engl. J. Med. 2018, 378, 1529–1536. [Google Scholar] [CrossRef] [PubMed]
- Al-Khan, A.; Youssef, Y.H.; Feldman, K.M.; Illsley, N.P.; Remache, Y.; Alvarez-Perez, J.; Mannion, C.; Alvarez, M.; Zamudio, S. Biomarkers of abnormally invasive placenta. Placenta 2020, 91, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Schwickert, A.; Chantraine, F.; Ehrlich, L.; Henrich, W.; Muallem, M.Z.; Nonnenmacher, A.; Petit, P.; Weizsäcker, K.; Braun, T. Maternal Serum VEGF Predicts Abnormally Invasive Placenta Better than NT-proBNP: A Multicenter Case-Control Study. Reprod. Sci. 2020, 28, 361–370. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Berkley, E.M.; Abuhamad, A. Imaging of Placenta Accreta Spectrum. Clin. Obstet. Gynecol. 2018, 61, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shan, R.; Song, Q.; Wei, X.; Liu, W.; Wang, G. Placenta percreta evaluated by MRI: Correlation with maternal morbidity. Arch. Gynecol. Obstet. 2020, 301, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Thiravit, S.; Lapatikarn, S.; Muangsomboon, K.; Suvannarerg, V.; Thiravit, P.; Korpraphong, P. MRI of placenta percreta: Differentiation from other entities of placental adhesive disorder. Radiol. Medica 2016, 122, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Al-Khan, A.; Gupta, V.; Illsley, N.P.; Mannion, C.; Koenig, C.; Bogomol, A.; Alvarez, M.; Zamudio, S. Maternal and fetal outcomes in placenta accreta after institution of team-managed care. Reprod. Sci. 2013, 21, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Kocherginsky, M.; Hibbard, J.U. Abnormal placentation: Twenty-year analysis. Am. J. Obstet. Gynecol. 2005, 192, 1458–1461. [Google Scholar] [CrossRef] [PubMed]
- Mehrabadi, A.; Hutcheon, J.A.; Liu, S.; Bartholomew, S.; Kramer, M.S.; Liston, R.M.; Joseph, K. Contribution of placenta accreta to the incidence of postpartum hemorrhage and severe postpartum hemorrhage. Obstet. Gynecol. 2015, 125, 814–821. [Google Scholar] [CrossRef] [PubMed]
- The American College of Obstetricians and Gynecologists. ACOG committee opinion no. 560: Medically indicated late-preterm and early-term deliveries. Obstet. Gynecol. 2013, 121, 908–910. [Google Scholar] [CrossRef] [PubMed]
- Robinson, B.K.; Grobman, W.A. Effectiveness of timing strategies for delivery of individuals with placenta previa and accreta. Obstet. Gynecol. 2010, 116, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Chantraine, F.; Blacher, S.; Berndt, S.; Palacios-Jaraquemada, J.; Sarioglu, N.; Nisolle, M.; Braun, T.; Munaut, C.; Foidart, J.M. Abnormal vascular architecture at the placental-maternal interface in placenta increta. Am. J. Obstet. Gynecol. 2012, 207, 188.e1–188.e9. [Google Scholar] [CrossRef] [PubMed]
- Biberoglu, E.; Kirbas, A.; Daglar, K.; Biberoglu, K.; Timur, H.; Demirtas, C.; Karabulut, E.; Danisman, N. Serum angiogenic profile in abnormal placentation. J. Matern. Neonatal Med. 2016, 29, 3193–3197. [Google Scholar] [CrossRef] [PubMed]
- Uyanıkoğlu, H.; İncebıyık, A.; Turp, A.B.; Çakmak, G.; Sak, S.; Hilali, N.G. Serum Angiogenic and Anti-angiogenic Markers in Pregnant Women with Placenta Percreta. Balk. Med. J. 2018, 35, 55–60. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Laban, M.; Ibrahim, E.A.; Elsafty, M.S.; Hassanin, A.S. Placenta accreta is associated with decreased decidual natural killer (dNK) cells population: A comparative pilot study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2014, 181, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Adler, E.; Madankumar, R.; Rosner, M.; Reznik, S.E. Increased placental trophoblast inclusions in placenta accreta. Placenta 2014, 35, 1075–1078. [Google Scholar] [CrossRef] [PubMed]
- McNally, L.; Zhou, Y.; Robinson, J.F.; Zhao, G.; Chen, L.M.; Chen, H.; Kim, M.Y.; Kapidzic, M.; Gormley, M.; Hannibal, R.; et al. Up-regulated cytotrophoblast DOCK4 contributes to over-invasion in placenta accreta spectrum. Proc. Natl. Acad. Sci. USA 2020, 117, 15852–15861. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Berezowsky, A.; Pardo, J.; Ben-Zion, M.; Wiznitzer, A.; Aviram, A. Second Trimester Biochemical Markers as Possible Predictors of Pathological Placentation: A Retrospective Case-Control Study. Fetal Diagn. Ther. 2019, 46, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Cantonwine, D.; Little, S.E.; McElrath, T.F.; Parry, S.I.; Lim, K.-H.; Wilkins-Haug, L.E. Angiogenic markers in pregnancies conceived through in vitro fertilization. Am. J. Obstet. Gynecol. 2015, 213, 212.e1–212.e8. [Google Scholar] [CrossRef] [PubMed]
- Büke, B.; Akkaya, H.; Demir, S.; Sağol, S.; Şimşek, D.; Başol, G.; Barutçuoğlu, B. Relationship between first trimester aneuploidy screening test serum analytes and placenta accreta. J. Matern. Neonatal Med. 2017, 31, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Samuel, A.; Bonanno, C.; Oliphant, A.; Batey, A.; Wright, J.D. Fraction of cell-free fetal DNA in the maternal serum as a predictor of abnormal placental invasion-a pilot study. Prenat. Diagn. 2013, 33, 1050–1053. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, J.; Saucier, J.B.; Feng, Y.; Jiang, Y.; Sinson, J.; McCombs, A.K.; Schmitt, E.S.; Peacock, S.; Chen, S.; et al. Non-invasive prenatal sequencing for multiple Mendelian monogenic disorders using circulating cell-free fetal DNA. Nat. Med. 2019, 25, 439–447, Erratum in Nat. Med. 2019, 25, 701–702. [Google Scholar] [CrossRef] [PubMed]
- Lyell, D.J.; Faucett, A.M.; Baer, R.J.; Blumenfeld, Y.J.; Druzin, M.L.; El-Sayed, Y.Y.; Shaw, G.M.; Currier, R.J.; Jelliffe-Pawlowski, L.L. Maternal serum markers, characteristics and morbidly adherent placenta in women with previa. Perinatol. 2015, 35, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Shainker, S.A.; Silver, R.M.; Modest, A.M.; Hacker, M.R.; Hecht, J.L.; Salahuddin, S.; Dillon, S.T.; Ciampa, E.J.; D’Alton, M.E.; Otu, H.H.; et al. Placenta accreta spectrum: Biomarker discovery using plasma proteomics. Am. J. Obstet. Gynecol. 2020, 223, 433.e1–433.e14. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Dai, L.; Chen, N.; Li, J.; Gao, Y.; Zhao, J.; Ding, L.; Xie, C.; Yi, X.; Deng, H.; et al. Integrative analysis provides multi-omics evidence for the pathogenesis of placenta percreta. J. Cell. Mol. Med. 2020, 24, 13837–13852. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gagnon, A.; Wilson, R.D.; Society of Obstetricians and Gynaecologists of Canada Genetics Committee. Obstetrical complications associated with abnormal maternal serum markers analytes. J. Obstet. Gynaecol. Can. 2008, 30, 918–932. [Google Scholar] [CrossRef] [PubMed]
- Uyanikoglu, H.; Sak, M.E.; Tatli, F.; Hilali, N.G.; Sak, S.; Incebiyik, A.; Barut, M.U.; Erel, O.; Gonel, A. Serum ischemia modified albumin level and its relationship with the thiol/disulfide balance in placenta percreta patients. J. Obstet. Gynaecol. 2018, 38, 1073–1077. [Google Scholar] [CrossRef] [PubMed]
- Ophir, E.; Tendler, R.; Odeh, M.; Khouri, S.; Oettinger, M. Creatine kinase as a biochemical marker in diagnosis of placenta increta and percreta. Am. J. Obstet. Gynecol. 1999, 180, 1039–1040. [Google Scholar] [CrossRef] [PubMed]
- Hilali, N.; Kocarslan, S.; Vural, M.; Incebiyik, A.; Camuzcuoglu, A.; Camuzcuoglu, H. Ki-67 proliferation index in patients with placenta previa percreta in the third trimester. Wien. Klin. Wochenschr. 2014, 127, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Pang, D.; Li, Y.; Zhou, J.; Liu, Y.; Yang, S.; Liang, K.; Yu, B. Serum miRNA biomarker discovery for placenta accreta spectrum. Placenta 2020, 101, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Mouillet, J.F.; Ouyang, Y.; Coyne, C.B.; Sadovsky, Y. MicroRNAs in placental health and disease. Am. J. Obstet. Gynecol. 2015, 213, S163–S172. [Google Scholar] [CrossRef]
- Lycoudi, A.; Mavreli, D.; Mavrou, A.; Papantoniou, N.; Kolialexi, A. miRNAs in pregnancy-related complications. Expert Rev. Mol. Diagn. 2015, 15, 999–1010. [Google Scholar] [CrossRef]
- Miura, K.; Miura, S.; Yamasaki, K.; Higashijima, A.; Kinoshita, A.; Yoshiura, K.; Masuzaki, H. Identification of pregnancy-associated microRNAs in maternal plasma. Clin. Chem. 2010, 56, 1767–1771. [Google Scholar] [CrossRef]
- Sedgwick, A.E.; D’Souza-Schorey, C. The biology of extracellular microvesicles. Traffic 2018, 19, 319–327. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tricarico, C.; Clancy, J.; D’Souza-Schorey, C. Biology and biogenesis of shed microvesicles. Small GTPases 2016, 8, 220–232. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mitchell, M.I.; Ma, J.; Carter, C.L.; Loudig, O. Circulating Exosome Cargoes Contain Functionally Diverse Cancer Biomarkers: From Biogenesis and Function to Purification and Potential Translational Utility. Cancers 2022, 14, 3350. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jin, J.; Menon, R. Placental exosomes: A proxy to understand pregnancy complications. Am. J. Reprod. Immunol. 2017, 79, e12788. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, A.; Nair, S.; Ormazabal, V.; Elfeky, O.; Garvey, C.E.; Longo, S.; Salomon, C. Circulating Placental Extracellular Vesicles and Their Potential Roles During Pregnancy. Ochsner J. 2020, 20, 439–445. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schuster, J.; Cheng, S.B.; Padbury, J.; Sharma, S. Placental extracellular vesicles and pre-eclampsia. Am. J. Reprod. Immunol. 2020, 85, e13297. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Miranda, J.; Paules, C.; Nair, S.; Lai, A.; Palma, C.; Scholz-Romero, K.; Rice, G.E.; Gratacos, E.; Crispi, F.; Salomon, C. Placental exosomes profile in maternal and fetal circulation in intrauterine growth restriction—Liquid biopsies to monitoring fetal growth. Placenta 2018, 64, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Salomon, C.; Torres, M.J.; Kobayashi, M.; Scholz-Romero, K.; Sobrevia, L.; Dobierzewska, A.; Illanes, S.E.; Mitchell, M.D.; Rice, G.E. A gestational profile of placental exosomes in maternal plasma and their effects on endothelial cell migration. PLoS ONE 2014, 9, e98667. [Google Scholar] [CrossRef] [PubMed]
- Karin-Kujundzic, V.; Sola, I.M.; Predavec, N.; Potkonjak, A.; Somen, E.; Mioc, P.; Serman, A.; Vranic, S.; Serman, L. Novel Epigenetic Biomarkers in Pregnancy-Related Disorders and Cancers. Cells 2019, 8, 1459. [Google Scholar] [CrossRef]
- Sussman, H.H.; Bowman, M.; Lewis, J.L., Jr. Placental alkaline phosphatase in maternal serum during normal and abnormal pregnancy. Nature 1968, 218, 359–360. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.; Scholz-Romero, K.; Perez, A.; Illanes, S.E.; Mitchell, M.D.; Rice, G.E.; Salomon, C. Placenta-derived exosomes continuously increase in maternal circulation over the first trimester of pregnancy. J. Transl. Med. 2014, 12, 204. [Google Scholar] [CrossRef] [PubMed]
- Czernek, L.; Düchler, M. Exosomes as Messengers Between Mother and Fetus in Pregnancy. Int. J. Mol. Sci. 2020, 21, 4264. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Williams, J.L.; Gatson, N.N.; Smith, K.M.; Almad, A.; McTigue, D.M.; Whitacre, C.C. Serum exosomes in pregnancy-associated immune modulation and neuroprotection during CNS autoimmunity. Clin. Immunol. 2013, 149, 236–243. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sabapatha, A.; Gercel-Taylor, C.; Taylor, D.D. Specific isolation of placenta-derived exosomes from the circulation of pregnant women and their immunoregulatory consequences. Am. J. Reprod. Immunol. 2006, 56, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.D.; Peiris, H.N.; Kobayashi, M.; Koh, Y.Q.; Duncombe, G.; Illanes, S.E.; Rice, G.E.; Salomon, C. Placental exosomes in normal and complicated pregnancy. Am. J. Obstet. Gynecol. 2015, 213 (Suppl. S4), S173–S181. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.I.; Ben-Dov, I.Z.; Liu, C.; Ye, K.; Chow, K.; Kramer, Y.; Gangadharan, A.; Park, S.; Fitzgerald, S.; Ramnauth, A.; et al. Extracellular Vesicle Capture by AnTibody of CHoice and Enzymatic Release (EV-CATCHER): A customizable purification assay designed for small-RNA biomarker identification and evaluation of circulating EVs. J. Extracell. Vesicles 2021, 10, e12110. [Google Scholar] [CrossRef] [PubMed]
- Holcar, M.; Ferdin, J.; Sitar, S.; Tušek-Žnidarič, M.; Dolžan, V.; Plemenitaš, A.; Žagar, E.; Lenassi, M. Enrichment of plasma extracellular vesicles for reliable quantification of their size and concentration for biomarker discovery. Sci. Rep. 2020, 10, 21346. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Silver, R.M.; Landon, M.B.; Rouse, D.J.; Leveno, K.J.; Spong, C.Y.; Thom, E.A.; Moawad, A.H.; Caritis, S.N.; Harper, M.; Wapner, R.J.; et al. Maternal morbidity associated with multiple repeat Cesarean deliveries. Obstet. Gynecol. 2006, 107, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Anderson-Bagga, F.M.; Sze, A. Placenta Previa. 2023 June 12. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar] [PubMed]
- Mitra, T.; Gulati, R.; Uppal, A.; Kumari, S.R.; Tripathy, S.; Ranjan, P.; Janardhanan, R. Prospecting of exosomal-miRNA signatures as prognostic marker for gestational diabetes mellitus and other adverse pregnancy outcomes. Front. Endocrinol. 2023, 14, 1097337. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Timofeeva, A.V.; Fedorov, I.S.; Suhova, Y.V.; Tarasova, A.M.; Ezhova, L.S.; Zabelina, T.M.; Vasilchenko, O.N.; Ivanets, T.Y.; Sukhikh, G.T. Diagnostic Role of Cell-Free miRNAs in Identifying Placenta Accreta Spectrum during First-Trimester Screening. Int. J. Mol. Sci. 2024, 25, 871. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Skalis, G.; Katsi, V.; Miliou, A.; Georgiopoulos, G.; Papazachou, O.; Vamvakou, G.; Nihoyannopoulos, P.; Tousoulis, D.; Makris, T. MicroRNAs in Preeclampsia. MicroRNA 2018, 8, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Zhou, T.; Yan, C.; Bao, J.; Yang, F.; Chao, S.; Zhou, M.; Xu, Z. A blood-based miRNA signature for early non-invasive diagnosis of preeclampsia. BMC Med. 2022, 20, 303. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cook, J.; Bennett, P.R.; Kim, S.H.; Teoh, T.G.; Sykes, L.; Kindinger, L.M.; Garrett, A.; Binkhamis, R.; MacIntyre, D.A.; Terzidou, V. First Trimester Circulating MicroRNA Biomarkers Predictive of Subsequent Preterm Delivery and Cervical Shortening. Sci. Rep. 2019, 9, 5861. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Illarionov, R.A.; Pachuliia, O.V.; Vashukova, E.S.; Tkachenko, A.A.; Maltseva, A.R.; Postnikova, T.B.; Nasykhova, Y.A.; Bespalova, O.N.; Glotov, A.S. Plasma miRNA Profile in High Risk of Preterm Birth during Early and Mid-Pregnancy. Genes 2022, 13, 2018. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ma, R.; Liang, Z.; Shi, X.; Xu, L.; Li, X.; Wu, J.; Zhao, L.; Liu, G. Exosomal miR-486-5p derived from human placental microvascular endothelial cells regulates proliferation and invasion of trophoblasts via targeting IGF1. Hum. Cell 2021, 34, 1310–1323. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Taga, S.; Hayashi, M.; Nunode, M.; Nakamura, N.; Ohmichi, M. miR-486-5p inhibits invasion and migration of HTR8/SVneo trophoblast cells by down-regulating ARHGAP5. Placenta 2022, 123, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.C.; Renthal, N.E.; Gerard, R.D.; Mendelson, C.R. The microRNA (miR)-199a/214 cluster mediates opposing effects of progesterone and estrogen on uterine contractility during pregnancy and labor. Mol. Endocrinol. 2012, 26, 1857–1867, Erratum in Mol. Endocrinol. 2013, 27, 188. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Peng, J.; Jiang, J.; Wang, H.; Feng, X.; Dong, X. miR-199a-3p suppresses cervical epithelial cell inflammation by inhibiting the HMGB1/TLR4/NF-κB pathway in preterm birth. Mol. Med. Rep. 2020, 22, 926–938. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mitchell, M.I.; Ben-Dov, I.Z.; Liu, C.; Wang, T.; Hazan, R.B.; Bauer, T.L.; Zakrzewski, J.; Donnelly, K.; Chow, K.; Ma, J.; et al. Non-invasive detection of orthotopic human lung tumors by microRNA expression profiling of mouse exhaled breath condensates and exhaled extracellular vesicles. Extracell. Vesicles Circ. Nucleic Acids 2024, 5, 138–164. [Google Scholar] [CrossRef]
- Fonseca, A.; Ayres de Campos, D. Maternal morbidity and mortality due to placenta accreta spectrum disorders. Best Pract. Res. Clin. Obstet. Gynaecol. 2020, 72, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Li, X.; Quan, X.; Yang, X.; Zheng, C.; Hao, X.; Qu, R.; Zhou, B. MiR-486 as an effective biomarker in cancer diagnosis and prognosis: A systematic review and meta-analysis. Oncotarget 2018, 9, 13948–13958. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, Y.; Fu, J.; Zhang, Z.; Qin, H. miR-486-5p regulates the migration and invasion of colorectal cancer cells through targeting PIK3R1. Oncol. Lett. 2018, 15, 7243–7248. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, L.; Mou, Y.P.; Wang, Y.Y.; Wang, H.J.; Mou, X.Z. miR-199a-3p targets ETNK1 to promote invasion and migration in gastric cancer cells and is associated with poor prognosis. Pathol.-Res. Pract. 2019, 215, 152511. [Google Scholar] [CrossRef] [PubMed]
- Faramin Lashkarian, M.; Hashemipour, N.; Niaraki, N.; Soghala, S.; Moradi, A.; Sarhangi, S.; Hatami, M.; Aghaei-Zarch, F.; Khosravifar, M.; Mohammadzadeh, A.; et al. MicroRNA-122 in human cancers: From mechanistic to clinical perspectives. Cancer Cell Int. 2023, 23, 29. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Al-Gazally, M.E.; Khan, R.; Imran, M.; Ramírez-Coronel, A.A.; Alshahrani, S.H.; Altalbawy, F.M.A.; Turki Jalil, A.; Romero-Parra, R.M.; Zabibah, R.S.; Shahid Iqbal, M.; et al. The role and mechanism of action of microRNA-122 in cancer: Focusing on the liver. Int. Immunopharmacol. 2023, 123, 110713. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ke, S.; Cheng, S.; Wu, T.; Yang, Y.; Liao, B. MicroRNA-151 regulates the growth, chemosensitivity and metastasis of human prostate cancer cells by targeting PI3K/AKT. J. BUON 2020, 25, 2045–2050. [Google Scholar] [PubMed]
- Qin, Y.; Liang, R.; Lu, P.; Lai, L.; Zhu, X. Depicting the Implication of miR-378a in Cancers. Technol. Cancer Res. Treat. 2022, 21, 15330338221134385. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, Z.; Xu, Y.; Wan, M.; Zeng, X.; Wu, J. miR-340: A multifunctional role in human malignant diseases. Int. J. Biol. Sci. 2021, 17, 236–246. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hu, L.; Ai, J.; Long, H.; Liu, W.; Wang, X.; Zuo, Y.; Li, Y.; Wu, Q.; Deng, Y. Integrative microRNA and gene profiling data analysis reveals novel biomarkers and mechanisms for lung cancer. Oncotarget 2016, 7, 8441–8454. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pisano, A.; Griñan-Lison, C.; Farace, C.; Fiorito, G.; Fenu, G.; Jiménez, G.; Scognamillo, F.; Peña-Martin, J.; Naccarati, A.; Pröll, J.; et al. The Inhibitory Role of miR-486-5p on CSC Phenotype Has Diagnostic and Prognostic Potential in Colorectal Cancer. Cancers 2020, 12, 3432. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tessema, M.; Yingling, C.M.; Picchi, M.A.; Wu, G.; Ryba, T.; Lin, Y.; Bungum, A.O.; Edell, E.S.; Spira, A.; Belinsky, S.A. ANK1 Methylation regulates expression of MicroRNA-486-5p and discriminates lung tumors by histology and smoking status. Cancer Lett. 2017, 410, 191–200. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Feng, L.; Jing, L.; Han, J.; Wang, G.; Liu, Y.; Zhang, X.; Wang, Y.; Wang, F.; Ma, H.; Liu, Y. MicroRNA 486-3p directly targets BIK and regulates apoptosis and invasion in colorectal cancer cells. OncoTargets Ther. 2018, 11, 8791–8801. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, X.P.; Hou, J.; Shen, X.Y.; Huang, C.Y.; Zhang, X.H.; Xie, Y.A.; Luo, X.L. MicroRNA-486-5p, which is downregulated in hepatocellular carcinoma, suppresses tumor growth by targeting PIK3R1. FEBS J. 2015, 282, 579–594. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Hou, G.; Xu, M.; Chen, M. Exosomal miR-122-3p represses the growth and metastasis of MCF-7/ADR cells by targeting GRK4-mediated activation of the Wnt/β-catenin pathway. Cell. Signal. 2024, 117, 111101. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Xu, M.; Yin, M.; Hong, J.; Chen, H.; Gao, Y.; Xie, C.; Shen, N.; Gu, S.; Mo, X. Exosomal hsa-miR199a-3p Promotes Proliferation and Migration in Neuroblastoma. Front. Oncol. 2019, 9, 459. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, J.; Ma, L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012, 31, 653–662. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pan, G.; Liu, Y.; Shang, L.; Zhou, F.; Yang, S. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun. 2021, 41, 199–217. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dong, B.; Li, S.; Zhu, S.; Yi, M.; Luo, S.; Wu, K. MiRNA-mediated EMT and CSCs in cancer chemoresistance. Exp. Hematol. Oncol. 2021, 10, 12. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dai, S.Y.; Kanenishi, K.; Ueno, M.; Sakamoto, H.; Hata, T. Hypoxia-inducible factor-2alpha is involved in enhanced apoptosis in the placenta from pregnancies with fetal growth restriction. Pathol. Int. 2004, 54, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Wang, Y.; Bai, S.; Xiao, Z.; Herva, R.; Piao, Y. Expression of integrins and extracellular matrix proteins at the maternal–fetal interface during tubal implantation. Reproduction 2003, 126, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; Zhu, L.; Li, L.; Kang, C. miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell. Mol. Biol. Lett. 2017, 22, 12. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, J.; Hyun, J.; Wang, S.; Lee, C.; Jung, Y. MicroRNA-378 is involved in hedgehog-driven epithelial-to-mesenchymal transition in hepatocytes of regenerating liver. Cell Death Dis. 2018, 9, 721. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cui, Z.; Sun, S.; Liu, Q.; Zhou, X.; Gao, S.; Peng, P.; Li, Q. MicroRNA-378-3p/5p suppresses the migration and invasiveness of oral squamous carcinoma cells by inhibiting KLK4 expression. Biochem. Cell Biol. 2020, 98, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Yu, X.; Xia, J.; Tang, X.; Tang, L.; Chen, F. MiR-486-3p targeting ECM1 represses cell proliferation and metastasis in cervical cancer. Biomed. Pharmacother. 2016, 80, 109–114. [Google Scholar] [CrossRef] [PubMed]
- ElKhouly, A.M.; Youness, R.A.; Gad, M.Z. MicroRNA-486-5p and microRNA-486-3p: Multifaceted pleiotropic mediators in oncological and non-oncological conditions. Non-Coding RNA Res. 2020, 5, 11–21, Erratum in Non-Coding RNA Res. 2020, 5, 219. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lin, X.M.; Wang, Z.J.; Lin, Y.X.; Chen, H. Decreased exosome-delivered miR-486-5p is responsible for the peritoneal metastasis of gastric cancer cells by promoting EMT progress. World J. Surg. Oncol. 2021, 19, 312. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yeh, T.C.; Huang, T.T.; Yeh, T.S.; Chen, Y.R.; Hsu, K.W.; Yin, P.H.; Lee, H.C.; Tseng, L.M. miR-151-3p Targets TWIST1 to Repress Migration of Human Breast Cancer Cells. PLoS ONE 2016, 11, e0168171. [Google Scholar] [PubMed] [PubMed Central]
- Daugaard, I.; Sanders, K.J.; Idica, A.; Vittayarukskul, K.; Hamdorf, M.; Krog, J.D.; Chow, R.; Jury, D.; Hansen, L.L.; Hager, H.; et al. miR-151a induces partial EMT by regulating E-cadherin in NSCLC cells. Oncogenesis 2017, 6, e366. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, B.; Xia, Y.; Lv, J.; Wang, W.; Xuan, Z.; Chen, C.; Jiang, T.; Fang, L.; Wang, L.; Li, Z.; et al. miR-151a-3p-rich small extracellular vesicles derived from gastric cancer accelerate liver metastasis via initiating a hepatic stemness-enhancing niche. Oncogene 2021, 40, 6180–6194. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Huang, Y.; Dong, S.; Qiao, C.; Yang, G.; Zhang, S.; Wang, C.; Xu, Y.; Zheng, F.; Yan, M. MiR-199a-3p/5p participated in TGF-β and EGF induced EMT by targeting DUSP5/MAP3K11 in pterygium. J. Transl. Med. 2020, 18, 332. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hou, L.K.; Yu, Y.; Xie, Y.G.; Wang, J.; Mao, J.F.; Zhang, B.; Wang, X.; Cao, X.C. miR-340 and ZEB1 negative feedback loop regulates TGF-β- mediated breast cancer progression. Oncotarget 2016, 7, 26016–26026. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wan, Y.; Huang, J.; Song, Y.; Gu, C.; Kong, J.; Zuo, L.; Chen, J. hsa-miR-340-5p inhibits epithelial-mesenchymal transition in endometriosis by targeting MAP3K2 and inactivating MAPK/ERK signaling. Open Med. 2022, 17, 566–576. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gong, H.; Lu, F.; Zeng, X.; Bai, Q. E2F transcription factor 1 (E2F1) enhances the proliferation, invasion and EMT of trophoblast cells by binding to Zinc Finger E-Box Binding Homeobox 1 (ZEB1). Bioengineered 2022, 13, 2360–2370. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mong, E.F.; Yang, Y.; Akat, K.M.; Canfield, J.; VanWye, J.; Lockhart, J.; Tsibris, J.C.M.; Schatz, F.; Lockwood, C.J.; Tuschl, T.; et al. Chromosome 19 microRNA cluster enhances cell reprogramming by inhibiting epithelial-to-mesenchymal transition. Sci. Rep. 2020, 10, 3029. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cindrova-Davies, T.; Yung, H.W.; Johns, J.; Spasic-Boskovic, O.; Korolchuk, S.; Jauniaux, E.; Burton, G.J.; Charnock-Jones, D.S. Oxidative stress, gene expression, and protein changes induced in the human placenta during labor. Am. J. Pathol. 2007, 171, 1168–1179. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Spillane, N.T.; Zamudio, S.; Alvarez-Perez, J.; Andrews, T.; Nyirenda, T.; Alvarez, M.; Al-Khan, A. Increased incidence of respiratory distress syndrome in neonates of mothers with abnormally invasive placentation. PLoS ONE 2018, 13, e0201266. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fenton, T.R.; Kim, J.H. A systematic review and meta-analysis to revise the Fenton growth chart for preterm infants. BMC Pediatr. 2013, 13, 59. [Google Scholar] [CrossRef] [PubMed Central]
- Loudig, O.; Liu, C.; Rohan, T.; Ben-Dov, I.Z. Retrospective MicroRNA Sequencing: Complementary DNA Library Preparation Protocol Using Formalin-fixed Paraffin embedded RNA Specimens. J. Vis. Exp. 2018, 135, 57471. [Google Scholar] [CrossRef] [PubMed]
- Loudig, O.; Wang, T.; Ye, K.; Lin, J.; Wang, Y.; Ramnauth, A.; Liu, C.; Stark, A.; Chitale, D.; Greenlee, R.; et al. Evaluation and Adaptation of a Laboratory-Based cDNA Library Preparation Protocol for Retrospective Sequencing of Archived MicroRNAs from up to 35-Year-Old Clinical FFPE Specimens. Int. J. Mol. Sci. 2017, 18, 627. [Google Scholar] [CrossRef] [PubMed]
- Mazeh, H.; Deutch, T.; Karas, A.; Bogardus, K.A.; Mizrahi, I.; Gur-Wahnon, D.; Ben-Dov, I.Z. Next-Generation Sequencing Identifies a Highly Accurate miRNA Panel That Distinguishes Well-Differentiated Thyroid Cancer from Benign Thyroid Nodules. Cancer Epidemiol. Biomark. Prev. 2018, 27, 858–863. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitchell, M.I.; Khalil, M.; Ben-Dov, I.Z.; Alverez-Perez, J.; Illsley, N.P.; Zamudio, S.; Al-Khan, A.; Loudig, O. Customizing EV-CATCHER to Purify Placental Extracellular Vesicles from Maternal Plasma to Detect Placental Pathologies. Int. J. Mol. Sci. 2024, 25, 5102. https://doi.org/10.3390/ijms25105102
Mitchell MI, Khalil M, Ben-Dov IZ, Alverez-Perez J, Illsley NP, Zamudio S, Al-Khan A, Loudig O. Customizing EV-CATCHER to Purify Placental Extracellular Vesicles from Maternal Plasma to Detect Placental Pathologies. International Journal of Molecular Sciences. 2024; 25(10):5102. https://doi.org/10.3390/ijms25105102
Chicago/Turabian StyleMitchell, Megan I., Marwa Khalil, Iddo Z. Ben-Dov, Jesus Alverez-Perez, Nicholas P. Illsley, Stacy Zamudio, Abdulla Al-Khan, and Olivier Loudig. 2024. "Customizing EV-CATCHER to Purify Placental Extracellular Vesicles from Maternal Plasma to Detect Placental Pathologies" International Journal of Molecular Sciences 25, no. 10: 5102. https://doi.org/10.3390/ijms25105102