Multi-Omics Characterization of Colon Mucosa and Submucosa/Wall from Crohn’s Disease Patients
Abstract
:1. Introduction
2. Results
2.1. Patient Demographics and Histologic Information
2.2. Comparison of Transcriptomics and Proteomics between Mucosa and Submucosa/Wall
2.3. WGCNA Reveals Compartment-Specific Regulation of Biological Functions
2.4. Comparison of Deconvoluted Cell Fractions between Mucosa and Submucosa/Wall
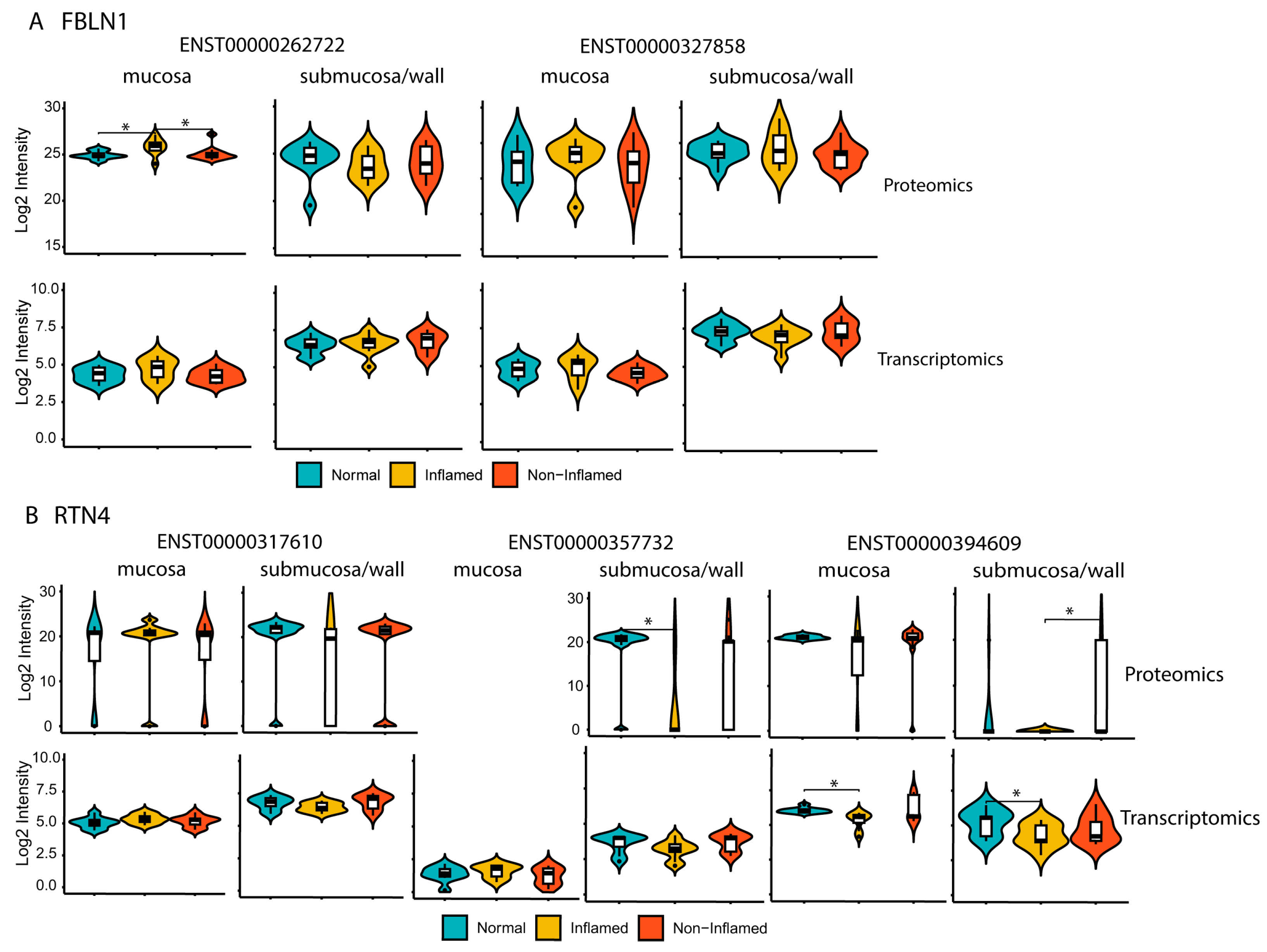
2.5. Differential Protein Isoforms between Mucosa and Submucosa/Wall Were Identified Using Proteogenomics
3. Discussion
4. Materials and Methods
4.1. Human Colon Resection Specimens and Dissection
4.2. Histology and Immunohistochemistry
4.3. RNA-Seq Library Preparation, Sequencing, Transcriptome Generation, and Analysis
4.4. Sample Preparation for Proteomics Analysis
4.5. Liquid Chromatography–Mass Spectrometry (LC-MS) Analysis
4.6. WGCNA Enrichment Analysis and Statistical Methods
4.7. Cell-Type Deconvolution and Statistical Methods
4.8. Proteogenomic Analysis to Identify Protein Isoforms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gajendran, M.; Loganathan, P.; Catinella, A.P.; Hashash, J.G. A Comprehensive Review and Update on Crohn’s Disease. Dis.-A-Mon. 2018, 64, 20–57. [Google Scholar] [CrossRef]
- Neurath, M.F.; Vieth, M. Different Levels of Healing in Inflammatory Bowel Diseases: Mucosal, Histological, Transmural, Barrier and Complete Healing. Gut 2023, 72, 2164–2183. [Google Scholar] [CrossRef]
- Agrawal, M.; Ebert, A.C.; Poulsen, G.; Ungaro, R.C.; Faye, A.S.; Jess, T.; Colombel, J.-F.; Allin, K.H. Early Ileocecal Resection for Crohn’s Disease Is Associated with Improved Long-Term Outcomes Compared with Anti-Tumor Necrosis Factor Therapy: A Population-Based Cohort Study. Gastroenterology 2023, 165, 976–985.e3. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Ricciuto, A.; Lewis, A.; D’Amico, F.; Dhaliwal, J.; Griffiths, A.M.; Bettenworth, D.; Sandborn, W.J.; Sands, B.E.; Reinisch, W.; et al. STRIDE-II: An Update on the Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE) Initiative of the International Organization for the Study of IBD (IOIBD): Determining Therapeutic Goals for Treat-to-Target Strategies in IBD. Gastroenterology 2021, 160, 1570–1583. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.R.; Bernardo, S.; Saraiva, S.; Gonçalves, A.R.; Santos, P.M.; Valente, A.; Correia, L.A.; Cortez-Pinto, H.; Magro, F. Proactive Infliximab Monitoring Improves the Rates of Transmural Remission in Crohn’s Disease: A Propensity Score–Matched Analysis. Inflamm. Bowel Dis. 2023, izad272. [Google Scholar] [CrossRef]
- Fernandes, S.R.; Bernardo, S.; Saraiva, S.; Gonçalves, A.R.; Santos, P.M.; Valente, A.; Correia, L.A.; Cortez-Pinto, H.; Magro, F. Tight Control Using Fecal Calprotectin and Early Disease Intervention Increase the Rates of Transmural Remission in Crohn’s Disease. United Eur. Gastroenterol. J. 2023, 12497. [Google Scholar] [CrossRef]
- Geyl, S.; Guillo, L.; Laurent, V.; D’Amico, F.; Danese, S.; Peyrin-Biroulet, L. Transmural Healing as a Therapeutic Goal in Crohn’s Disease: A Systematic Review. Lancet Gastroenterol. Hepatol. 2021, 6, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.Y.; Kim, E.S.; Jeon, T.Y.; Lee, Y.M.; Lee, S.M.; Choe, B.; Choe, Y.H.; Kang, B. Transmural Healing Evaluated by Magnetic Resonance Enterography in Paediatric Patients with Crohn’s Disease Receiving Maintenance Treatment with Biologics. Aliment. Pharmacol. Ther. 2022, 56, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Dolinger, M.T.; Calabrese, E.; Pizzolante, F.; Abreu, M.T. Current and Novel Uses of Intestinal Ultrasound in Inflammatory Bowel Disease. Gastroenterol. Hepatol. 2023, 19, 447–457. [Google Scholar]
- Ma, L.; Li, W.; Zhuang, N.; Yang, H.; Liu, W.; Zhou, W.; Jiang, Y.; Li, J.; Zhu, Q.; Qian, J. Comparison of Transmural Healing and Mucosal Healing as Predictors of Positive Long-Term Outcomes in Crohn’s Disease. Ther. Adv. Gastroenterol. 2021, 14, 17562848211016260. [Google Scholar] [CrossRef]
- Takenaka, K.; Kawamoto, A.; Kitazume, Y.; Fujii, T.; Udagawa, Y.; Shimizu, H.; Hibiya, S.; Nagahori, M.; Ohtsuka, K.; Watanabe, M.; et al. Transmural Remission Characterized by High Biologic Concentrations Demonstrates Better Prognosis in Crohn’s Disease. J. Crohn’s Colitis 2022, 17, 855–862. [Google Scholar] [CrossRef] [PubMed]
- de Lange, K.M.; Moutsianas, L.; Lee, J.C.; Lamb, C.A.; Luo, Y.; Kennedy, N.A.; Jostins, L.; Rice, D.L.; Gutierrez-Achury, J.; Ji, S.-G.; et al. Genome-Wide Association Study Implicates Immune Activation of Multiple Integrin Genes in Inflammatory Bowel Disease. Nat. Genet. 2017, 49, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Momozawa, Y.; Dmitrieva, J.; Théâtre, E.; Deffontaine, V.; Rahmouni, S.; Charloteaux, B.; Crins, F.; Docampo, E.; Elansary, M.; Gori, A.-S.; et al. IBD Risk Loci Are Enriched in Multigenic Regulatory Modules Encompassing Putative Causative Genes. Nat. Commun. 2018, 9, 2427. [Google Scholar] [CrossRef] [PubMed]
- Acera, M.G.; Patankar, J.V.; Diemand, L.; Siegmund, B.; Neurath, M.F.; Wirtz, S.; Becker, C. Comparative Transcriptomics of IBD Patients Indicates Induction of Type 2 Immunity Irrespective of the Disease Ideotype. Front. Med. 2021, 8, 664045. [Google Scholar] [CrossRef]
- Eshelman, M.A.; Harris, L.; Deiling, S.; Koltun, W.A.; Jeganathan, N.A.; Yochum, G.S. Transcriptomic Analysis of Ileal Tissue from Crohn’s Disease Patients Identifies Extracellular Matrix Genes That Distinguish Individuals by Age at Diagnosis. Physiol. Genom. 2020, 52, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Assadsangabi, A.; Evans, C.A.; Corfe, B.M.; Lobo, A. Application of Proteomics to Inflammatory Bowel Disease Research: Current Status and Future Perspectives. Gastroenterol. Res. Pr. 2019, 2019, 1426954. [Google Scholar] [CrossRef] [PubMed]
- Aldars-García, L.; Gisbert, J.P.; Chaparro, M. Metabolomics Insights into Inflammatory Bowel Disease: A Comprehensive Review. Pharmaceuticals 2021, 14, 1190. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, M.; Allin, K.H.; Petralia, F.; Colombel, J.-F.; Jess, T. Multiomics to Elucidate Inflammatory Bowel Disease Risk Factors and Pathways. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 399–409. [Google Scholar] [CrossRef]
- Sudhakar, P.; Alsoud, D.; Wellens, J.; Verstockt, S.; Arnauts, K.; Verstockt, B.; Vermeire, S. Tailoring Multi-Omics to Inflammatory Bowel Diseases: All for One and One for All. J. Crohn’s Colitis 2022, 16, 1306–1320. [Google Scholar] [CrossRef]
- Jin, L.; Li, L.; Hu, C.; Paez-Cortez, J.; Bi, Y.; Macoritto, M.; Cao, S.; Tian, Y. Integrative Analysis of Transcriptomic and Proteomic Profiling in Inflammatory Bowel Disease Colon Biopsies. Inflamm. Bowel Dis. 2019, 25, 1906–1918. [Google Scholar] [CrossRef]
- Menden, K.; Marouf, M.; Oller, S.; Dalmia, A.; Magruder, D.S.; Kloiber, K.; Heutink, P.; Bonn, S. Deep Learning–Based Cell Composition Analysis from Tissue Expression Profiles. Sci. Adv. 2020, 6, eaba2619. [Google Scholar] [CrossRef] [PubMed]
- Smillie, C.S.; Biton, M.; Ordovas-Montanes, J.; Sullivan, K.M.; Burgin, G.; Graham, D.B.; Herbst, R.H.; Rogel, N.; Slyper, M.; Waldman, J.; et al. Intra- and Inter-Cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 2019, 178, 714–730.e22. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.W.; Hill, D.G.; Jones, S.A. Understanding Immune Cells in Tertiary Lymphoid Organ Development: It Is All Starting to Come Together. Front. Immunol. 2016, 7, 401. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, B. CustomProDB: An R Package to Generate Customized Protein Databases from RNA-Seq Data for Proteomics Search. Bioinformatics 2013, 29, 3235–3237. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Cooley, M.A.; Jarnicki, A.G.; Hsu, A.C.-Y.; Nair, P.M.; Haw, T.J.; Fricker, M.; Gellatly, S.L.; Kim, R.Y.; Inman, M.D.; et al. Fibulin-1 Regulates the Pathogenesis of Tissue Remodeling in Respiratory Diseases. JCI Insight 2016, 1, e86380. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lu, R.; Shu, L.; Chen, Y.; Zhao, J.; Dai, J.; Huang, Q.; Li, X.; Meng, W.; Long, F.; et al. An Integrated Map of Fibroblastic Populations in Human Colon Mucosa and Cancer Tissues. Commun. Biol. 2022, 5, 1326. [Google Scholar] [CrossRef] [PubMed]
- Janker, L.; Schuster, D.; Bortel, P.; Hagn, G.; Meier-Menches, S.M.; Mohr, T.; Mader, J.C.; Slany, A.; Bileck, A.; Brunmair, J.; et al. Multiomics-Empowered Deep Phenotyping of Ulcerative Colitis Identifies Biomarker Signatures Reporting Functional Remission States. J. Crohn’s Colitis 2023, 17, 1514–1527. [Google Scholar] [CrossRef] [PubMed]
- Kinchen, J.; Chen, H.H.; Parikh, K.; Antanaviciute, A.; Jagielowicz, M.; Fawkner-Corbett, D.; Ashley, N.; Cubitt, L.; Mellado-Gomez, E.; Attar, M.; et al. Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell 2018, 175, 372–386.e17. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.S.; Strittmatter, S.M. The Reticulons: A Family of Proteins with Diverse Functions. Genome Biol. 2007, 8, 234. [Google Scholar] [CrossRef]
- Nagarajan, U.M.; Lochamy, J.; Chen, X.; Beresford, G.W.; Nilsen, R.; Jensen, P.E.; Boss, J.M. Class II Transactivator Is Required for Maximal Expression of HLA-DOB in B Cells. J. Immunol. 2002, 168, 1780–1786. [Google Scholar] [CrossRef]
- Lin, A.; Yan, W.-H. The Emerging Roles of Human Leukocyte Antigen-F in Immune Modulation and Viral Infection. Front. Immunol. 2019, 10, 964. [Google Scholar] [CrossRef] [PubMed]
- Castro-Dopico, T.; Clatworthy, M.R. IgG and Fcγ Receptors in Intestinal Immunity and Inflammation. Front. Immunol. 2019, 10, 805. [Google Scholar] [CrossRef] [PubMed]
- Wiendl, M.; Becker, E.; Müller, T.M.; Voskens, C.J.; Neurath, M.F.; Zundler, S. Targeting Immune Cell Trafficking—Insights From Research Models and Implications for Future IBD Therapy. Front. Immunol. 2021, 12, 656452. [Google Scholar] [CrossRef]
- Gu, P.; Theiss, A.; Han, J.; Feagins, L.A. Increased Cell Adhesion Molecules, PECAM-1, ICAM-3, or VCAM-1, Predict Increased Risk for Flare in Patients with Quiescent Inflammatory Bowel Disease. J. Clin. Gastroenterol. 2017, 51, 522–527. [Google Scholar] [CrossRef]
- Murahashi, M.; Kogami, A.; Muramoto, A.; Hoshino, H.; Akama, T.O.; Mitoma, J.; Goi, T.; Hirayama, A.; Okamura, T.; Nagaya, T.; et al. Vascular E-Selectin Expression Detected in Formalin-Fixed, Paraffin-Embedded Sections with an E-Selectin Monoclonal Antibody Correlates with Ulcerative Colitis Activity. J. Histochem. Cytochem. 2022, 70, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, M.; Pohin, M.; Jackson, M.A.; Korsunsky, I.; Bullers, S.J.; Rue-Albrecht, K.; Christoforidou, Z.; Sathananthan, D.; Thomas, T.; Ravindran, R.; et al. IL-1-Driven Stromal–Neutrophil Interactions Define a Subset of Patients with Inflammatory Bowel Disease That Does Not Respond to Therapies. Nat. Med. 2021, 27, 1970–1981. [Google Scholar] [CrossRef] [PubMed]
- Juzenas, S.; Hübenthal, M.; Lindqvist, C.M.; Kruse, R.; Steiert, T.A.; Degenhardt, F.; Schulte, D.; Nikolaus, S.; Zeissig, S.; Bergemalm, D.; et al. Detailed Transcriptional Landscape of Peripheral Blood Points to Increased Neutrophil Activation in Treatment-Naïve Inflammatory Bowel Disease. J. Crohn’s Colitis 2022, 16, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X.; Wang, S.; Ba, X.; Xu, T.; Wang, X.; Zeng, X. RhoGDI2 Positively Regulates the Rho GTPases Activation in Response to the Β2 Outside-in Signaling in T Cells Adhesion and Migration on ICAM-1. J. Leukoc. Biol. 2019, 106, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Kardol-Hoefnagel, T.; van Logtestijn, S.A.L.M.; Otten, H.G. A Review on the Function and Regulation of ARHGDIB/RhoGDI2 Expression Including the Hypothetical Role of ARHGDIB/RhoGDI2 Autoantibodies in Kidney Transplantation. Transplant. Direct 2020, 6, e548. [Google Scholar] [CrossRef] [PubMed]
- Heybeli, C. The Complement System and Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2016, 22, E22. [Google Scholar] [CrossRef]
- Fleming, A.; Castro-Dopico, T.; Clatworthy, M.R. B Cell Class Switching in Intestinal Immunity in Health and Disease. Scand. J. Immunol. 2022, 95, e13139. [Google Scholar] [CrossRef] [PubMed]
- Targan, S.R.; Landers, C.J.; Yang, H.; Lodes, M.J.; Cong, Y.; Papadakis, K.A.; Vasiliauskas, E.; Elson, C.O.; Hershberg, R.M. Antibodies to CBir1 Flagellin Define a Unique Response That Is Associated Independently with Complicated Crohn’s Disease. Gastroenterology 2005, 128, 2020–2028. [Google Scholar] [CrossRef]
- Spencer, J.; Bemark, M. Human Intestinal B Cells in Inflammatory Diseases. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Elson, C.O.; Dunkin, D. Epicutaneous Immunotherapy with CBir1 Alleviates Intestinal Inflammation. Inflamm. Bowel Dis. 2023, 29, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Ho, G.; Theiss, A.L. Mitochondria and Inflammatory Bowel Diseases: Toward a Stratified Therapeutic Intervention. Annu. Rev. Physiol. 2021, 84, 435–459. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Zan, X.; Jia, Z.; Zheng, L.; Gu, Y.; Liu, F.; Han, Y.; Xu, C.; Wu, A.; Zhi, Q. Crosstalk between Alternative Splicing and Inflammatory Bowel Disease: Basic Mechanisms, Biotechnological Progresses and Future Perspectives. Clin. Transl. Med. 2023, 13, e1479. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Liu, Y.; Yi, X.; Olsen, L.K.; Zhu, H.; Gao, Q.; Zhou, H.; Zhang, B. SEPepQuant Enhances the Detection of Possible Isoform Regulations in Shotgun Proteomics. Nat. Commun. 2023, 14, 5809. [Google Scholar] [CrossRef] [PubMed]
- Fancello, L.; Burger, T. An Analysis of Proteogenomics and How and When Transcriptome-Informed Reduction of Protein Databases Can Enhance Eukaryotic Proteomics. Genome Biol. 2022, 23, 132. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential Analyses for RNA-Seq: Transcript-Level Estimates Improve Gene-Level Inferences. F1000Research 2016, 4, 1521. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Stekhoven, D.J.; Bühlmann, P. MissForest—Non-Parametric Missing Value Imputation for Mixed-Type Data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Bi, Y.; Hu, C.; Qu, J.; Shen, S.; Wang, X.; Tian, Y. A Comparative Study of Evaluating Missing Value Imputation Methods in Label-Free Proteomics. Sci. Rep. 2021, 11, 1760. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Wang, J.; Macoritto, M.; Guay, H.; Davis, J.W.; Levesque, M.C.; Cao, X. The Clinical Response of Upadacitinib and Risankizumab Is Associated with Reduced Inflammatory Bowel Disease Anti-TNF-α Inadequate Response Mechanisms. Inflamm. Bowel Dis. 2022, 29, 771–782. [Google Scholar] [CrossRef]






| Colon Mucosa | Colon Submucosa/Wall | |||||
|---|---|---|---|---|---|---|
| Non-IBD Control (Normal) | CD Patient, Inflamed (CD) | CD Patient, Non-Inflamed (CD-NI) | Non-IBD Control (Normal) | CD Patient, Inflamed (CD) | CD Patient, Non-Inflamed (CD-NI) | |
| Total number | 8 | 8 | 9 | 9 | 10 | 8 |
| Gender (Male/Female) * | 4/4 | 3/5 | 4/5 | 3/6 | 4/6 | 3/5 |
| Age (years) † | 68 (56–78) | 43.5 (33–70) | 46 (33–70) | 69 (56–78) | 46 (26–70) | 42.5 (26–70) |
| Race (Caucasian/African American/NA) | 6/1/1 | 8/0/0 | 8/0/1 | 8/0/1 | 9/1/0 | 7/1/0 |
| Smoking history (yes/no/NA) | 2/5/1 | 3/5/0 | 4/5/0 | 2/6/1 | 4/6/0 | 3/5/0 |
| Alcohol drinking history (yes/no/NA) | 1/1/6 | 3/5/0 | 3/6/0 | 2/1/6 | 1/5/4 | 2/2/4 |
| Biologics treatment (yes/no/NA) | 0/1/7 | 1/5/2 | 1/6/2 | 0/2/7 | 1/4/5 | 0/4/4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, L.; Macoritto, M.; Wang, J.; Bi, Y.; Wang, F.; Suarez-Fueyo, A.; Paez-Cortez, J.; Hu, C.; Knight, H.; Mascanfroni, I.; et al. Multi-Omics Characterization of Colon Mucosa and Submucosa/Wall from Crohn’s Disease Patients. Int. J. Mol. Sci. 2024, 25, 5108. https://doi.org/10.3390/ijms25105108
Jin L, Macoritto M, Wang J, Bi Y, Wang F, Suarez-Fueyo A, Paez-Cortez J, Hu C, Knight H, Mascanfroni I, et al. Multi-Omics Characterization of Colon Mucosa and Submucosa/Wall from Crohn’s Disease Patients. International Journal of Molecular Sciences. 2024; 25(10):5108. https://doi.org/10.3390/ijms25105108
Chicago/Turabian StyleJin, Liang, Michael Macoritto, Jing Wang, Yingtao Bi, Fei Wang, Abel Suarez-Fueyo, Jesus Paez-Cortez, Chenqi Hu, Heather Knight, Ivan Mascanfroni, and et al. 2024. "Multi-Omics Characterization of Colon Mucosa and Submucosa/Wall from Crohn’s Disease Patients" International Journal of Molecular Sciences 25, no. 10: 5108. https://doi.org/10.3390/ijms25105108





