A Workflow for Meaningful Interpretation of Classification Results from Handheld Ambient Mass Spectrometry Analysis Probes
Abstract
:1. Introduction
2. Experimental Design
2.1. Prospective, Retrospective Approaches and Appropriate Patient Inclusion/Exclusion Criteria
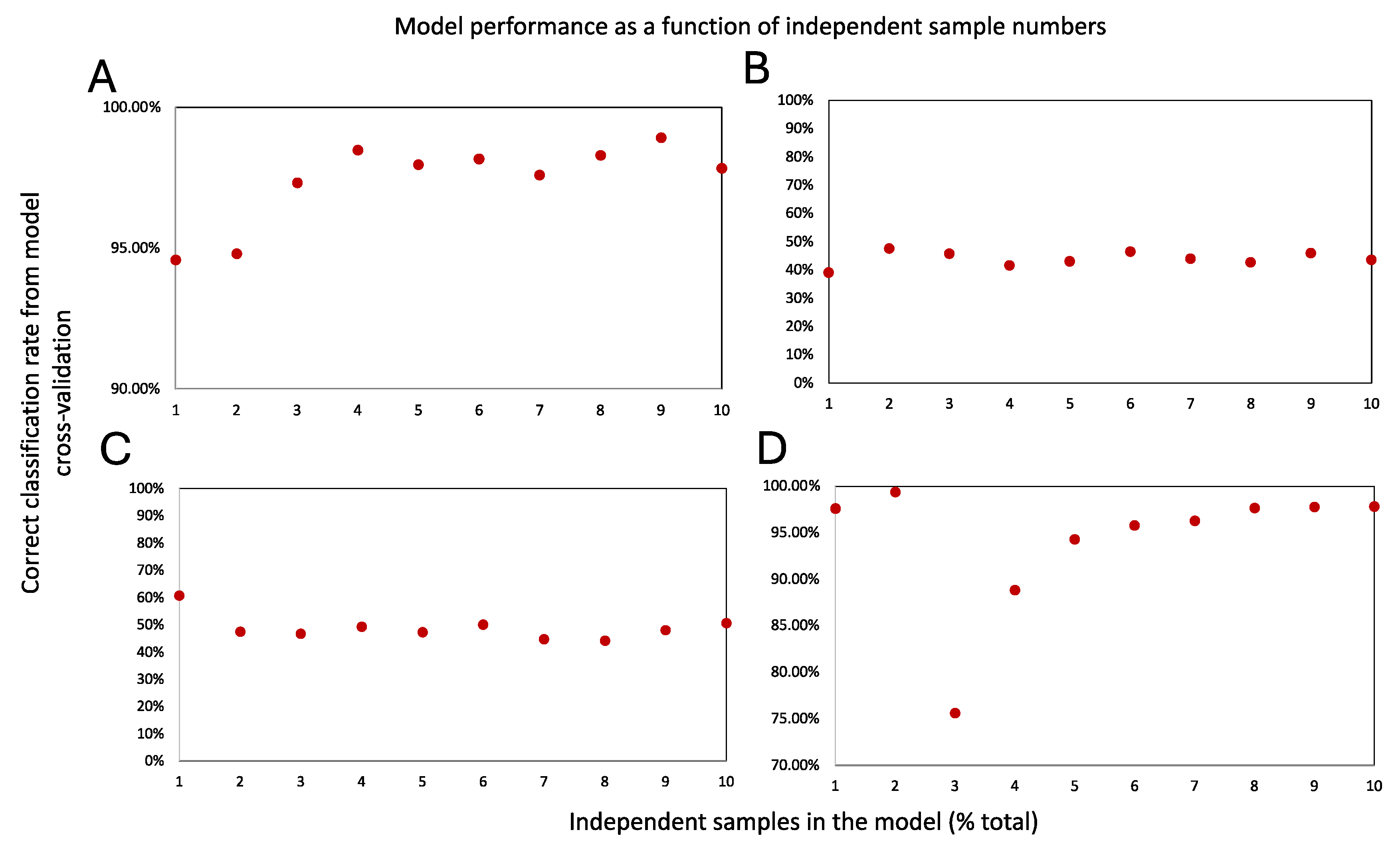
2.2. Suitable Study Size and Data Acquisition Strategy
3. Creation and Validation of Mass Spectrometry Molecular Models Using Intelligent Data Analysis Tools
3.1. Data Analysis Approaches and Preprocessing of Data
3.2. Data Analysis: First Steps and Approach Planning Considerations
3.3. Assessment of Molecular Model’s Robustness
4. Interpretation of Data
4.1. The Need to Understand the Classifiers
4.2. Revising the Model Based on Rationalizable Classifiers and Further Validating It
5. Caveats and Additional Points to Consider
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Feider, C.L.; Krieger, A.; DeHoog, R.J.; Eberlin, L.S. Ambient Ionization Mass Spectrometry: Recent Developments and Applications. Anal. Chem. 2019, 91, 4266–4290. [Google Scholar] [CrossRef]
- Zhang, J.; Sans, M.; Garza, K.Y.; Eberlin, L.S. Mass Spectrometry Technologies to Advance Care for Cancer Patients in Clinical and Intraoperative Use. Mass Spectrom. Rev. 2020, 40, 692–720. [Google Scholar] [CrossRef]
- Zhang, J.; Rector, J.; Lin, J.Q.; Young, J.H.; Sans, M.; Katta, N.; Giese, N.; Yu, W.; Nagi, C.; Suliburk, J.; et al. Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system. Sci. Transl. Med. 2017, 9, eaan3968. [Google Scholar] [CrossRef] [PubMed]
- Takats, Z.; Strittmatter, N.; McKenzie, J.S. Ambient Mass Spectrometry in Cancer Research. Adv. Cancer Res. 2017, 134, 231–256. [Google Scholar]
- Ogrinc, N.; Saudemont, P.; Takats, Z.; Salzet, M.; Fournier, I. Cancer Surgery 2.0: Guidance by Real-Time Molecular Technologies. Trends Mol. Med. 2021, 27, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Sachfer, K.C.; Szaniszlo, T.; Gunther, S.; Balog, J.; Denes, J.; Keseru, M.; Dezso, B.; Toth, M.; Spengler, B.; Takats, Z. In situ, real-time identification of biological tissues by ultraviolet and infrared laser desorption ionization mass spectrometry. Anal. Chem. 2011, 83, 1632–1640. [Google Scholar] [CrossRef]
- Balog, J.; Sasi-Szabo, L.; Kinross, J.; Lewis, M.R.; Muirhead, L.J.; Veselkov, K.; Mirnezami, R.; Dezso, B.; Damjanovich, L.; Darzi, A.; et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci. Transl. Med. 2013, 5, 194ra93. [Google Scholar] [CrossRef]
- Paraskevaidi, M.; Cameron, S.J.S.; Whelan, E.; Bowden, S.; Tzafetas, M.; Mitra, A.; Semertzidou, A.; Athanasiou, A.; Bennett, P.R.; MacIntyre, D.A.; et al. Laser-assisted rapid evaporative ionisation mass spectrometry (LA-REIMS) as a metabolomics platform in cervical cancer screening. EBioMedicine 2020, 60, 103017. [Google Scholar] [CrossRef]
- Schafer, K.C.; Balog, J.; Szaniszlo, T.; Szalay, D.; Mezey, G.; Denes, J.; Bognar, L.; Oertel, M.; Takats, Z. Real time analysis of brain tissue by direct combination of ultrasonic surgical aspiration and sonic spray mass spectrometry. Anal. Chem. 2011, 83, 7729–7735. [Google Scholar] [CrossRef] [PubMed]
- Saudemont, P.; Quanico, J.; Robin, Y.M.; Baud, A.; Balog, J.; Fatou, B.; Tierny, D.; Pascal, Q.; Minier, K.; Pottier, M.; et al. Real-Time Molecular Diagnosis of Tumors Using Water-Assisted Laser Desorption/Ionization Mass Spectrometry Technology. Cancer Cell 2018, 34, 840–851.e4. [Google Scholar] [CrossRef]
- Woolman, M.; Kuzan-Fischer, C.M.; Ferry, I.; Kiyota, T.; Luu, B.; Wu, M.; Munoz, D.G.; Das, S.; Aman, A.; Taylor, M.D.; et al. Picosecond Infrared Laser Desorption Mass Spectrometry Identifies Medulloblastoma Subgroups on Intrasurgical Timescales. Cancer Res. 2019, 79, 2426–2434. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Woolman, M.; Kiyota, T.; Pires, L.; Zaidi, M.; Hofer, S.O.P.; Leong, W.; Wouters, B.G.; Ghazarian, D.; Chan, A.W.; et al. Picosecond Infrared Laser Mass Spectrometry Identifies a Metabolite Array for 10 s Diagnosis of Select Skin Cancer Types: A Proof-of-Concept Feasibility Study. Anal. Chem. 2022, 94, 16821–16830. [Google Scholar] [CrossRef] [PubMed]
- Woolman, M.; Katz, L.; Tata, A.; Basu, S.S.; Zarrine-Afsar, A. Breaking Through the Barrier: Regulatory Considerations Relevant to Ambient Mass Spectrometry at the Bedside. Clin. Lab. Med. 2021, 41, 221–246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sans, M.; DeHoog, R.J.; Garza, K.Y.; King, M.E.; Feider, C.L.; Bensussan, A.; Keating, M.F.; Lin, J.Q.; Povilaitis, S.C.; et al. Clinical Translation and Evaluation of a Handheld and Biocompatible Mass Spectrometry Probe for Surgical Use. Clin. Chem. 2021, 67, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Sans, M.; Zhang, J.; Lin, J.Q.; Feider, C.L.; Giese, N.; Breen, M.T.; Sebastian, K.; Liu, J.; Sood, A.K.; Eberlin, L.S. Performance of the MasSpec Pen for Rapid Diagnosis of Ovarian Cancer. Clin. Chem. 2019, 65, 674–683. [Google Scholar] [CrossRef]
- Kuo, T.H.; Dutkiewicz, E.P.; Pei, J.; Hsu, C.C. Ambient Ionization Mass Spectrometry Today and Tomorrow: Embracing Challenges and Opportunities. Anal. Chem. 2020, 92, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Rankin-Turner, S.; Reynolds, J.C.; Turner, M.A.; Heaney, L.M. Applications of ambient ionization mass spectrometry in 2021: An annual review. Analytical Science Advances 2022, 3, 67–89. [Google Scholar] [CrossRef]
- Brown, H.M.; Pirro, V.; Cooks, R.G. From DESI to the MasSpec Pen: Ambient Ionization Mass Spectrometry for Tissue Analysis and Intrasurgical Cancer Diagnosis. Clin. Chem. 2018, 64, 628–630. [Google Scholar] [CrossRef]
- Katz, L.; Tata, A.; Woolman, M.; Zarrine-Afsar, A. Lipid Profiling in Cancer Diagnosis with Hand-Held Ambient Mass Spectrometry Probes: Addressing the Late-Stage Performance Concerns. Metabolites 2021, 11, 660. [Google Scholar] [CrossRef]
- USP Pharmacopeial Convention. Guidance on developing and validating nontargeted methods for adulteration detection. In Appendix XVIII; US Pharmacopoeia: Rockville, MD, USA, 2016. [Google Scholar]
- Available online: https://www.fda.gov/media/89841/download (accessed on 12 December 2023).
- Available online: https://www.accessdata.fda.gov/cdrh_docs/reviews/K130831.pdf (accessed on 12 December 2023).
- Available online: https://www.accessdata.fda.gov/cdrh_docs/reviews/K162950.pdf (accessed on 12 December 2023).
- Ioannidis, J.P.A.; Bossuyt, P.M.M. Waste, Leaks, and Failures in the Biomarker Pipeline. Clin. Chem. 2017, 63, 963–972. [Google Scholar] [CrossRef]
- Katz, L.; Woolman, M.; Tata, A.; Zarrine-Afsar, A. Potential impact of tissue molecular heterogeneity on ambient mass spectrometry profiles: A note of caution in choosing the right disease model. Anal. Bioanal. Chem. 2021, 413, 2655–2664. [Google Scholar] [CrossRef]
- Woolman, M.; Ferry, I.; Kuzan-Fischer, C.M.; Wu, M.; Zou, J.; Kiyota, T.; Isik, S.; Dara, D.; Aman, A.; Das, S.; et al. Rapid determination of medulloblastoma subgroup affiliation with mass spectrometry using a handheld picosecond infrared laser desorption probe. Chem. Sci. 2017, 8, 6508–6519. [Google Scholar] [CrossRef]
- Woolman, M.; Zarrine-Afsar, A. Platforms for rapid cancer characterization by ambient mass spectrometry: Advancements, challenges and opportunities for improvement towards intrasurgical use. Analyst 2018, 143, 2717–2722. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Beroukhim, R.; Golub, T.R. Genomic evolution of cancer models: Perils and opportunities. Nat. Rev. Cancer 2019, 19, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Alewijn, M.; van der Voet, H.; van Ruth, S. Validation of multivariate classification methods using analytical fingerprints–concept and case study on organic feed for laying hens. J. Food Compos. Anal. 2016, 51, 15–23. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; van Velzen, E.J.J.; van Duijnhoven, J.P.M.; van Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2013, 1, 92–107. [Google Scholar]
- Liebal, U.W.; Phan, A.N.T.; Sudhakar, M.; Raman, K.; Blank, L.M. Machine Learning Applications for Mass Spectrometry-Based Metabolomics. Metabolites 2020, 10, 243. [Google Scholar] [CrossRef] [PubMed]
- D'Amico, E.J.; Neilands, T.B.; Zambarano, R. Power analysis for multivariate and repeated measures designs: A flexible approach using the SPSS MANOVA procedure. Behav. Res. Methods Instrum. Comput. 2001, 33, 479–484. [Google Scholar] [CrossRef]
- Blaise, B.J.; Correia, G.; Tin, A.; Young, J.H.; Vergnaud, A.C.; Lewis, M.; Pearce, J.T.; Elliott, P.; Nicholson, J.K.; Holmes, E.; et al. Power Analysis and Sample Size Determination in Metabolic Phenotyping. Anal. Chem. 2016, 88, 5179–5188. [Google Scholar] [CrossRef]
- Blaise, B.J. Data-driven sample size determination for metabolic phenotyping studies. Anal. Chem. 2013, 85, 8943–8950. [Google Scholar] [CrossRef]
- Dudzik, D.; Barbas-Bernardos, C.; Garcia, A.; Barbas, C. Quality assurance procedures for mass spectrometry untargeted metabolomics. a review. J. Pharm. Biomed. Anal. 2018, 147, 149–173. [Google Scholar] [CrossRef]
- Ali, N.; Girnus, S.; Rosch, P.; Popp, J.; Bocklitz, T. Sample-Size Planning for Multivariate Data: A Raman-Spectroscopy-Based Example. Anal. Chem. 2018, 90, 12485–12492. [Google Scholar] [CrossRef]
- Katz, L.; Woolman, M.; Talbot, F.; Amara-Belgadi, S.; Wu, M.; Tortorella, S.; Das, S.; Ginsberg, H.J.; Zarrine-Afsar, A. Dual Laser and Desorption Electrospray Ionization Mass Spectrometry Imaging Using the Same Interface. Anal. Chem. 2020, 92, 6349–6357. [Google Scholar] [CrossRef]
- Porcari, A.M.; Zhang, J.; Garza, K.Y.; Rodrigues-Peres, R.M.; Lin, J.Q.; Young, J.H.; Tibshirani, R.; Nagi, C.; Paiva, G.R.; Carter, S.A.; et al. Multicenter Study Using Desorption-Electrospray-Ionization-Mass-Spectrometry Imaging for Breast-Cancer Diagnosis. Anal. Chem. 2018, 90, 11324–11332. [Google Scholar] [CrossRef]
- McGrath, T.F.; Haughey, S.A.; Patterson, J.; Fauhl-Hassek, C.; Donarski, J.; Alewijn, M.; van Ruth, S.; Elliott, C.T. What are the scientific challenges in moving from targeted to non-targeted methods for food fraud testing and how can they be addressed?—Spectroscopy case study. Trends Food Sci. Technol. 2018, 76, 38–55. [Google Scholar] [CrossRef]
- Ghazalpour, A.; Bennett, B.; Petyuk, V.A.; Orozco, L.; Hagopian, R.; Mungrue, I.N.; Farber, C.R.; Sinsheimer, J.; Kang, H.M.; Furlotte, N.; et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011, 7, e1001393. [Google Scholar] [CrossRef] [PubMed]
- Morrissy, A.S.; Cavalli, F.M.G.; Remke, M.; Ramaswamy, V.; Shih, D.J.H.; Holgado, B.L.; Farooq, H.; Donovan, L.K.; Garzia, L.; Agnihotri, S.; et al. Spatial heterogeneity in medulloblastoma. Nat. Genet. 2017, 49, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Peck, B.; Schulze, A. Lipid Metabolism at the Nexus of Diet and Tumor Microenvironment. Trends Cancer 2019, 5, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Dahdah, N.; Gonzalez-Franquesa, A.; Samino, S.; Gama-Perez, P.; Herrero, L.; Perales, J.C.; Yanes, O.; Malagon, M.D.M.; Garcia-Roves, P.M. Effects of Lifestyle Intervention in Tissue-Specific Lipidomic Profile of Formerly Obese Mice. Int. J. Mol. Sci. 2021, 22, 3694. [Google Scholar] [CrossRef] [PubMed]
- Krauss, R.M.; Burkman, R.T., Jr. The metabolic impact of oral contraceptives. Am. J. Obstet. Gynecol. 1992, 167 Pt 2, 1177–1184. [Google Scholar] [CrossRef]
- Katz, L.; Kiyota, T.; Woolman, M.; Wu, M.; Pires, L.; Fiorante, A.; Ye, L.A.; Leong, W.; Berman, H.K.; Ghazarian, D.; et al. Metabolic Lipids in Melanoma Enable Rapid Determination of Actionable BRAF-V600E Mutation with Picosecond Infrared Laser Mass Spectrometry in 10 s. Anal. Chem. 2023, 95, 14430–14439. [Google Scholar] [CrossRef]
- King, M.E.; Zhang, J.; Lin, J.Q.; Garza, K.Y.; DeHoog, R.J.; Feider, C.L.; Bensussan, A.; Sans, M.; Krieger, A.; Badal, S.; et al. Rapid diagnosis and tumor margin assessment during pancreatic cancer surgery with the MasSpec Pen technology. Proc. Natl. Acad. Sci. USA 2021, 118, e2104411118. [Google Scholar] [CrossRef]
- St John, E.R.; Balog, J.; McKenzie, J.S.; Rossi, M.; Covington, A.; Muirhead, L.; Bodai, Z.; Rosini, F.; Speller, A.V.M.; Shousha, S.; et al. Rapid evaporative ionisation mass spectrometry of electrosurgical vapours for the identification of breast pathology: Towards an intelligent knife for breast cancer surgery. Breast Cancer Res. 2017, 19, 59. [Google Scholar] [CrossRef]
- Povilaitis, S.C.; Chakraborty, A.; Kirkpatrick, L.M.; Downey, R.D.; Hauger, S.B.; Eberlin, L.S. Identifying Clinically Relevant Bacteria Directly from Culture and Clinical Samples with a Handheld Mass Spectrometry Probe. Clin. Chem. 2022, 68, 1459–1470. [Google Scholar] [CrossRef]
- Keating, M.F.; Zhang, J.; Feider, C.L.; Retailleau, S.; Reid, R.; Antaris, A.; Hart, B.; Tan, G.; Milner, T.E.; Miller, K.; et al. Integrating the MasSpec Pen to the da Vinci Surgical System for In Vivo Tissue Analysis during a Robotic Assisted Porcine Surgery. Anal. Chem. 2020, 92, 11535–11542. [Google Scholar] [CrossRef]
- Djulbegovic, B.; Hozo, I.; Mayrhofer, T.; van den Ende, J.; Guyatt, G. The threshold model revisited. J. Eval. Clin. Pract. 2019, 25, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Cahan, A.; Gilon, D.; Manor, O.; Paltiel, O. Probabilistic reasoning and clinical decision-making: Do doctors overestimate diagnostic probabilities? QJM 2003, 96, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Habbema, J.D. Clinical decision theory: The threshold concept. Neth. J. Med. 1995, 47, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Dill, A.L.; Eberlin, L.S.; Costa, A.B.; Ifa, D.R.; Cooks, R.G. Data quality in tissue analysis using desorption electrospray ionization. Anal. Bioanal. Chem. 2011, 401, 1949–1961. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.E.; Scott, D.R. Optimization and testing of mass spectral library search algorithms for compound identification. J. Am. Soc. Mass Spectrom. 1994, 5, 859–866. [Google Scholar] [CrossRef]
- Eberlin, L.S.; Tibshirani, R.J.; Zhang, J.; Longacre, T.A.; Berry, G.J.; Bingham, D.B.; Norton, J.A.; Zare, R.N.; Poultsides, G.A. Molecular assessment of surgical-resection margins of gastric cancer by mass-spectrometric imaging. Proc. Natl. Acad. Sci. USA 2014, 111, 2436–2441. [Google Scholar] [CrossRef]
- Sans, M.; Gharpure, K.; Tibshirani, R.; Zhang, J.; Liang, L.; Liu, J.; Young, J.H.; Dood, R.L.; Sood, A.K.; Eberlin, L.S. Metabolic Markers and Statistical Prediction of Serous Ovarian Cancer Aggressiveness by Ambient Ionization Mass Spectrometry Imaging. Cancer Res. 2017, 77, 2903–2913. [Google Scholar] [CrossRef]
- Calligaris, D.; Norton, I.; Feldman, D.R.; Ide, J.L.; Dunn, I.F.; Eberlin, L.S.; Cooks, R.G.; Jolesz, F.A.; Golby, A.J.; Santagata, S.; et al. Mass spectrometry imaging as a tool for surgical decision-making. J. Mass Spectrom. 2013, 48, 1178–1187. [Google Scholar] [CrossRef]
- Santagata, S.; Eberlin, L.S.; Norton, I.; Calligaris, D.; Feldman, D.R.; Ide, J.L.; Liu, X.; Wiley, J.S.; Vestal, M.L.; Ramkissoon, S.H.; et al. Intraoperative mass spectrometry mapping of an onco-metabolite to guide brain tumor surgery. Proc. Natl. Acad. Sci. USA 2014, 111, 11121–11126. [Google Scholar] [CrossRef]
- Eberlin, L.S. DESI-MS imaging of lipids and metabolites from biological samples. Methods Mol. Biol. 2014, 1198, 299–311. [Google Scholar]
- Eberlin, L.S.; Norton, I.; Orringer, D.; Dunn, I.F.; Liu, X.; Ide, J.L.; Jarmusch, A.K.; Ligon, K.L.; Jolesz, F.A.; Golby, A.J.; et al. Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc. Natl. Acad. Sci. USA 2013, 110, 1611–1616. [Google Scholar] [CrossRef] [PubMed]
- Ifa, D.R.; Eberlin, L.S. Ambient Ionization Mass Spectrometry for Cancer Diagnosis and Surgical Margin Evaluation. Clin. Chem. 2016, 62, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Fatou, B.; Saudemont, P.; Leblanc, E.; Vinatier, D.; Mesdag, V.; Wisztorski, M.; Focsa, C.; Salzet, M.; Ziskind, M.; Fournier, I. In vivo Real-Time Mass Spectrometry for Guided Surgery Application. Sci. Rep. 2016, 6, 25919. [Google Scholar] [CrossRef] [PubMed]
- Seddiki, K.; Saudemont, P.; Precioso, F.; Ogrinc, N.; Wisztorski, M.; Salzet, M.; Fournier, I.; Droit, A. Cumulative learning enables convolutional neural network representations for small mass spectrometry data classification. Nat. Commun. 2020, 11, 5595. [Google Scholar] [CrossRef] [PubMed]
- DeHoog, R.J.; King, M.E.; Keating, M.F.; Zhang, J.; Sans, M.; Feider, C.L.; Garza, K.Y.; Bensussan, A.; Krieger, A.; Lin, J.Q.; et al. Intraoperative Identification of Thyroid and Parathyroid Tissues During Human Endocrine Surgery Using the MasSpec Pen. JAMA Surg. 2023, 158, 1050–1059. [Google Scholar] [CrossRef]
- Handley, K.F.; Sims, T.T.; Bateman, N.W.; Glassman, D.; Foster, K.I.; Lee, S.; Yao, J.; Yao, H.; Fellman, B.M.; Liu, J.; et al. Classification of High-Grade Serous Ovarian Cancer Using Tumor Morphologic Characteristics. JAMA Netw. Open 2022, 5, e2236626. [Google Scholar] [CrossRef]
- Lee, E.S.; Durant, T.J.S. Supervised machine learning in the mass spectrometry laboratory: A tutorial. J. Mass Spectrom. Adv. Clin. Lab. 2022, 23, 1–6. [Google Scholar] [CrossRef]
- Greenacre, M.; Groenen, P.J.F.; Hastie, T.; D’Enza, A.I.; Markos, A.; Tuzhilina, E. Principal component analysis. Nat. Rev. Methods Primers 2022, 2, 100. [Google Scholar] [CrossRef]
- Bhamre, T.; Zhao, Z.; Singer, A. Mahalanobis Distance for Class Averaging of Cryo-Em Images. arXiv 2016, arXiv:1611.03193v4. [Google Scholar]
- Bodai, Z.; Cameron, S.; Bolt, F.; Simon, D.; Schaffer, R.; Karancsi, T.; Balog, J.; Rickards, T.; Burke, A.; Hardiman, K.; et al. Effect of Electrode Geometry on the Classification Performance of Rapid Evaporative Ionization Mass Spectrometric (REIMS) Bacterial Identification. J. Am. Soc. Mass Spectrom. 2018, 29, 26–33. [Google Scholar] [CrossRef]
- Yao, L.; Lin, T.B. Evolutionary Mahalanobis Distance-Based Oversampling for Multi-Class Imbalanced Data Classification. Sensors 2021, 21, 6616. [Google Scholar] [CrossRef] [PubMed]
- Sylvain, A.; Alain, C. A survey of cross-validation procedures for model selection. Stat. Surv. 2010, 4, 40–79. [Google Scholar]
- Little, M.A.; Varoquaux, G.; Saeb, S.; Lonini, L.; Jayaraman, A.; Mohr, D.C.; Kording, K.P. Using and understanding cross-validation strategies. Perspectives on Saeb et al. Gigascience 2017, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hollon, T.C.; Lewis, S.; Pandian, B.; Niknafs, Y.S.; Garrard, M.R.; Garton, H.; Maher, C.O.; McFadden, K.; Snuderl, M.; Lieberman, A.P.; et al. Rapid Intraoperative Diagnosis of Pediatric Brain Tumors Using Stimulated Raman Histology. Cancer Res. 2018, 78, 278–289. [Google Scholar] [CrossRef]
- Li, M.; Gao, Q.; Yu, T. Kappa statistic considerations in evaluating inter-rater reliability between two raters: Which, when and context matters. BMC Cancer 2023, 23, 799. [Google Scholar] [CrossRef]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. (Zagreb) 2012, 22, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, H.S.; Yoon, Y.C.; Cha, M.J.; Lee, S.H.; Kim, E.S. Differentiating between spinal schwannomas and meningiomas using MRI: A focus on cystic change. PLoS ONE 2020, 15, e0233623. [Google Scholar] [CrossRef] [PubMed]
- Woolman, M.; Qiu, J.; Kuzan-Fischer, C.M.; Ferry, I.; Dara, D.; Katz, L.; Daud, F.; Wu, M.; Ventura, M.; Bernards, N.; et al. In situ tissue pathology from spatially encoded mass spectrometry classifiers visualized in real time through augmented reality. Chem. Sci. 2020, 11, 8723–8735. [Google Scholar] [CrossRef] [PubMed]
- Brereton, R.G. Consequences of sample size, variable selection, and model validation and optimisation, for predicting classification ability from analytical data. TrAC Trends Anal. Chem. 2006, 25, 1103–1111. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Kjeldahl, K.; Bro, R. Some common misunderstandings in chemometrics. J. Chemom. 2010, 24, 558–564. [Google Scholar] [CrossRef]
- Paschali, M.; Conjeti, S.; Navarro, F.; Navab, N. Generalizability vs. Robustness: Investigating Medical Imaging Networks Using Adversarial Examples. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018, Proceedings of the 21st International Conference, Granada, Spain, 16–20 September 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 493–501. [Google Scholar]
- Patocka, J.; Hon, Z. Ethylene glycol, hazardous substance in the household. Acta Medica (Hradec Kralove) 2010, 53, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.; Schreiner, P.; Leung, A.; Stainton, P.; Reid, S.; Yaworski, E.; Lutwyche, P.; Heyes, J. Optimizing Lipid Nanoparticles for Delivery in Primates. Adv. Mater. 2023, 35, e2211420. [Google Scholar] [CrossRef]
- Giakoumatos, E.C.; Gascoigne, L.; Gumi-Audenis, B.; Garcia, A.G.; Tuinier, R.; Voets, I.K. Impact of poly(ethylene glycol) functionalized lipids on ordering and fluidity of colloid supported lipid bilayers. Soft Matter 2022, 18, 7569–7578. [Google Scholar] [CrossRef]
- Boger, D.L.; Henriksen, S.J.; Cravatt, B.F. Oleamide: An endogenous sleep-inducing lipid and prototypical member of a new class of biological signaling molecules. Curr. Pharm. Des. 1998, 4, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, A.; Degenhardt, O.S.; McDonald, G.R.; Narang, D.; Paulsen, I.M.; Kozuska, J.L.; Holt, A. On the disruption of biochemical and biological assays by chemicals leaching from disposable laboratory plasticware. Can. J. Physiol. Pharmacol. 2012, 90, 697–703. [Google Scholar] [CrossRef]
- Butovich, I.A. On the lipid composition of human meibum and tears: Comparative analysis of nonpolar lipids. Investig. Ophthalmol. Vis. Sci. 2008, 49, 3779–3789. [Google Scholar] [CrossRef] [PubMed]
- Bhunia, K.; Sablani, S.S.; Tang, J.; Rasco, B. Migration of Chemical Compounds from Packaging Polymers during Microwave, Conventional Heat Treatment, and Storage. Compr. Rev. Food Sci. Food Saf. 2013, 12, 523–545. [Google Scholar] [CrossRef]
- Naumoska, K.; Jug, U.; Metličar, V.; Vovk, I. Oleamide, a Bioactive Compound, Unwittingly Introduced into the Human Body through Some Plastic Food/Beverages and Medicine Containers. Foods 2020, 9, 549. [Google Scholar] [CrossRef]
- Koo, Y.P.; Yahaya, N.; Omar, W.A.W. Analysis of dibutyl phthalate and oleamide in stingless bee honey harvested from plastic cups. Sains Malays. 2017, 46, 449–455. [Google Scholar] [CrossRef]
- Thongkhao, K.; Prombutara, P.; Phadungcharoen, T.; Wiwatcharakornkul, W.; Tungphatthong, C.; Sukrong, M.; Sukrong, S. Integrative approaches for unmasking hidden species in herbal dietary supplement products: What is in the capsule? J. Food Compos. Anal. 2020, 93, 103616. [Google Scholar] [CrossRef]
- Niziol, J.; Ossolinski, K.; Plaza-Altamer, A.; Kolodziej, A.; Ossolinska, A.; Ossolinski, T.; Nieczaj, A.; Ruman, T. Untargeted urinary metabolomics for bladder cancer biomarker screening with ultrahigh-resolution mass spectrometry. Sci. Rep. 2023, 13, 9802. [Google Scholar] [CrossRef]
- Chen, J.; Hou, H.; Chen, H.; Luo, Y.; Zhang, L.; Zhang, Y.; Liu, H.; Zhang, F.; Liu, Y.; Wang, A.; et al. Urinary metabolomics for discovering metabolic biomarkers of laryngeal cancer using UPLC-QTOF/MS. J. Pharm. Biomed. Anal. 2019, 167, 83–89. [Google Scholar] [CrossRef]
- Arendowski, A.; Ossolinski, K.; Niziol, J.; Ruman, T. Screening of Urinary Renal Cancer Metabolic Biomarkers with Gold Nanoparticles-assisted Laser Desorption/Ionization Mass Spectrometry. Anal. Sci. 2020, 36, 1521–1525. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Xie, G.; Jia, W. Metabonomics of human colorectal cancer: New approaches for early diagnosis and biomarker discovery. J. Proteome Res. 2014, 13, 3857–3870. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007, 35, W606–W612. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Tanimu, J.J.; Hassan, M.; Kakudi, H.A.; Robert, P. Evaluation of Recursive Feature Elimination and LASSO Regularization-based optimized feature selection approaches for cervical cancer prediction. In Proceedings of the 2021 IEEE 14th International Symposium on Embedded Multicore/Many-core Systems-on-Chip (MCSoC), Singapore, 20–23 December 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 333–339. [Google Scholar]
- Peter, K.T.; Phillips, A.L.; Knolhoff, A.M.; Gardinali, P.R.; Manzano, C.A.; Miller, K.E.; Pristner, M.; Sabourin, L.; Sumarah, M.W.; Warth, B.; et al. Nontargeted Analysis Study Reporting Tool: A Framework to Improve Research Transparency and Reproducibility. Anal. Chem. 2021, 93, 13870–13879. [Google Scholar] [CrossRef] [PubMed]
- Smets, T.; Verbeeck, N.; Claesen, M.; Asperger, A.; Griffioen, G.; Tousseyn, T.; Waelput, W.; Waelkens, E.; De Moor, B. Evaluation of Distance Metrics and Spatial Autocorrelation in Uniform Manifold Approximation and Projection Applied to Mass Spectrometry Imaging Data. Anal. Chem. 2019, 91, 5706–5714. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiorante, A.; Ye, L.A.; Tata, A.; Kiyota, T.; Woolman, M.; Talbot, F.; Farahmand, Y.; Vlaminck, D.; Katz, L.; Massaro, A.; et al. A Workflow for Meaningful Interpretation of Classification Results from Handheld Ambient Mass Spectrometry Analysis Probes. Int. J. Mol. Sci. 2024, 25, 3491. https://doi.org/10.3390/ijms25063491
Fiorante A, Ye LA, Tata A, Kiyota T, Woolman M, Talbot F, Farahmand Y, Vlaminck D, Katz L, Massaro A, et al. A Workflow for Meaningful Interpretation of Classification Results from Handheld Ambient Mass Spectrometry Analysis Probes. International Journal of Molecular Sciences. 2024; 25(6):3491. https://doi.org/10.3390/ijms25063491
Chicago/Turabian StyleFiorante, Alexa, Lan Anna Ye, Alessandra Tata, Taira Kiyota, Michael Woolman, Francis Talbot, Yasamine Farahmand, Darah Vlaminck, Lauren Katz, Andrea Massaro, and et al. 2024. "A Workflow for Meaningful Interpretation of Classification Results from Handheld Ambient Mass Spectrometry Analysis Probes" International Journal of Molecular Sciences 25, no. 6: 3491. https://doi.org/10.3390/ijms25063491




