Polycations as Aptamer-Binding Modulators for Sensitive Fluorescence Anisotropy Assay of Aflatoxin B1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Sample Preparation
2.2. Synthesis of the Fluorescein-Labeled Derivative of Aflatoxin B1 (AFB1-EDF)
2.3. The Characterization of the Interactions between Aptamer Variants and AFB1-EDF Using Fluorescence Anisotropy
2.4. The Determination of Dissociating Constants Using Fluorescence Anisotropy Measurements
2.5. The Characterization of Influence of Different Medium Compounds or Co-Solvents on the Fluorescence Anisotropy
- (1)
- The 5-fold dilutions from 10 to 0.08 μM of poly-L-lysine, 30 kDa;
- (2)
- The 5-fold dilutions from 8750 to 70 μM of polyethylene glycol, 8 kDa;
- (3)
- The 3-fold dilutions of MgAcetate2 from 1 M to 37 mM in a magnesium-free TB.
2.6. The FA-Based Detection of Aflatoxin B1
2.7. Sample Preparation
3. Results and Discussion
3.1. The Scheme of the Proposed Fluorescence Anisotropy Assay for Aflatoxin B1 Using Polycation Modulators
3.2. The Influence of the Length of the Aptamer Stem Region on the Binding of Labeled AFB1
3.3. The Influence of the Length of the Aptamer Stem Region on the Binding of Labeled AFB1
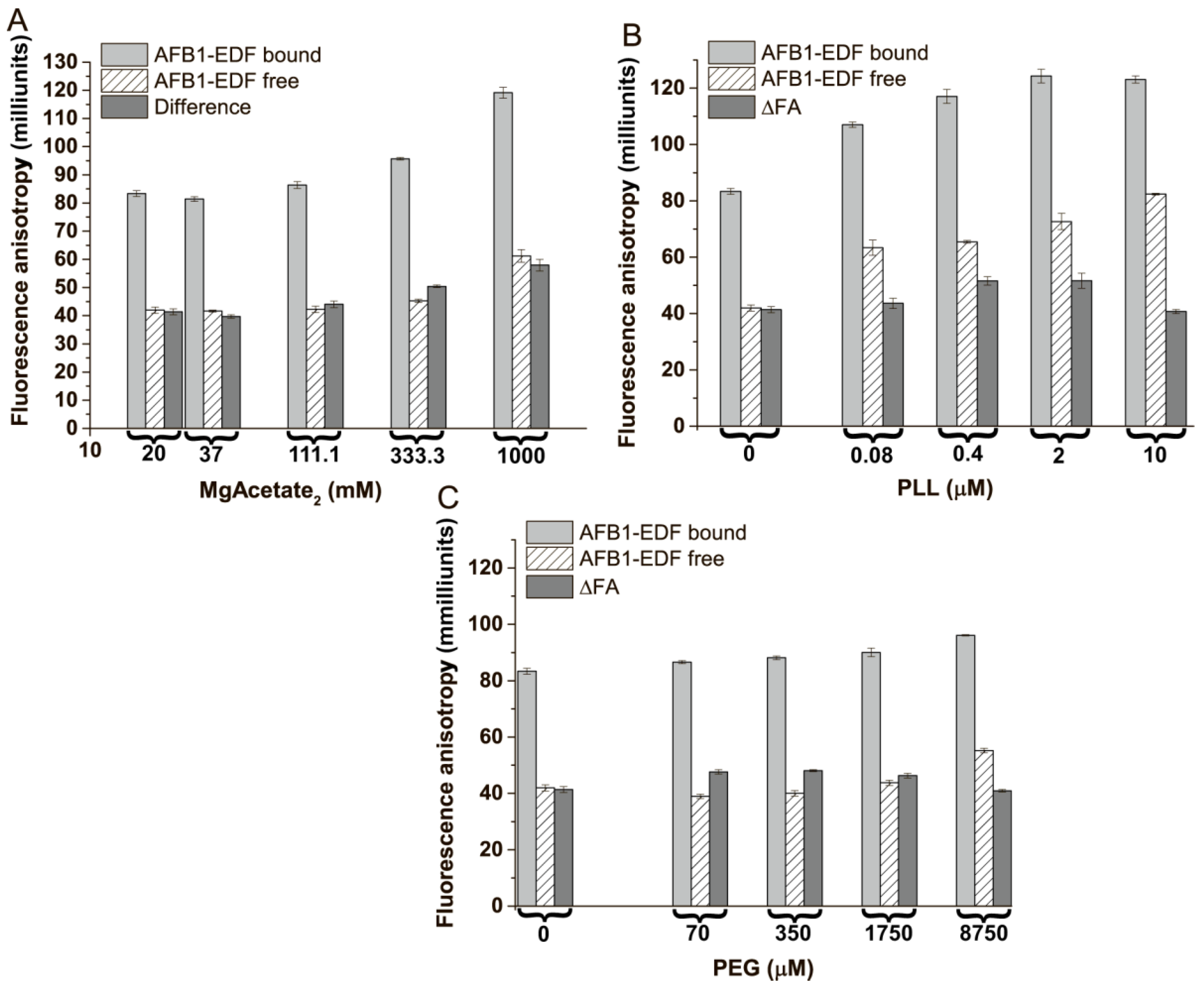
3.4. The Influence of Polymers and Cations on Fluorescence Anisotropy of Fluorescein-Labeled AFB1 in Free and Bound State
3.5. The Influence of Different Co-Solvents on Fluorescence Anisotropy of Fluorescein-Labeled AFB1 in the Free and Bound State
3.6. The Influence of the Aptamer Complexation with Streptavidin on Fluorescence Anisotropy
3.7. The Cooperative Influence of Polycations and PEG on the Difference in FA between the Bound and Free States of Labeled AFB1
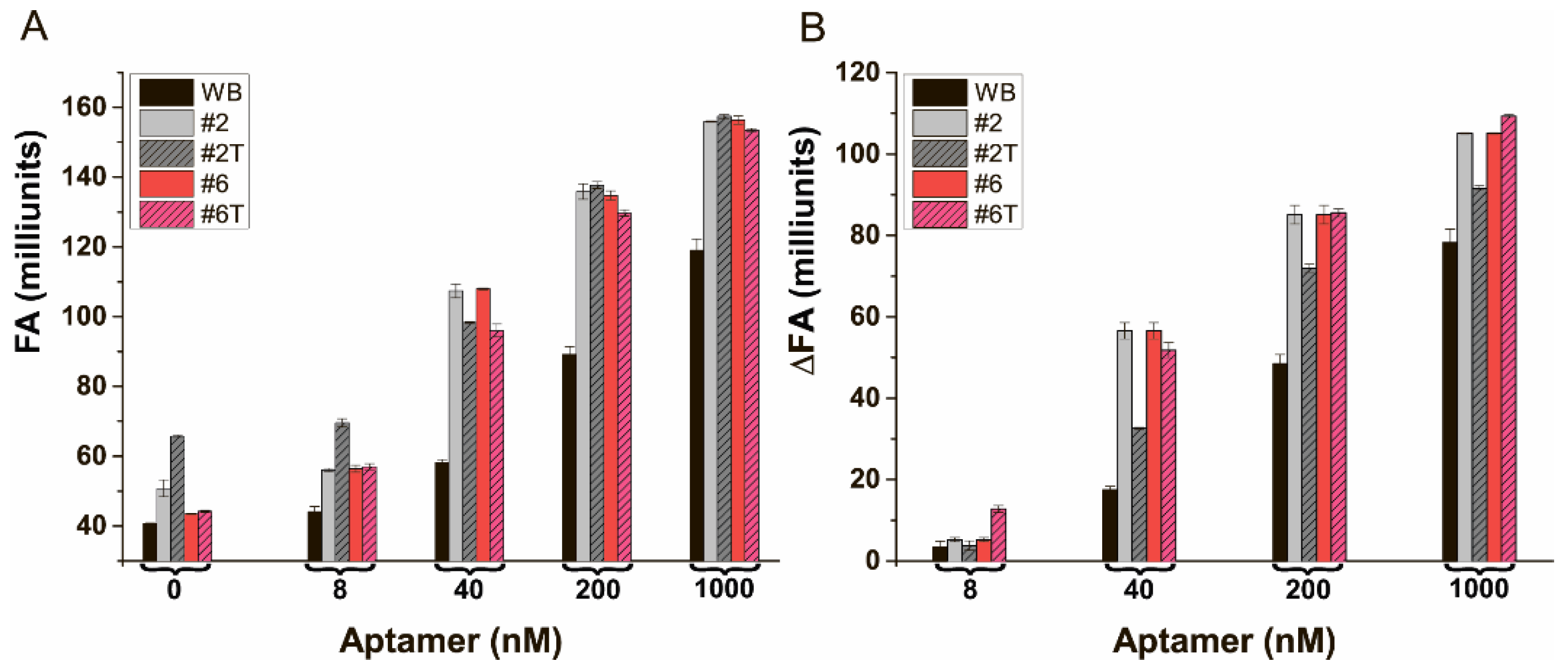
3.8. The Characterization of FA of the Bound and Free State of AFB1-EDF at Different Concentrations of the Aptamer in the Selected Media
3.9. The Comparison of FA-Based Aptamer Assays of AFB1 in TB and Buffer #6
3.10. Detection of AFB1 in Food Samples
3.11. Comparison of the Developed FA Aptamer Assay with Other Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hendrickson, O.D.; Taranova, N.A.; Zherdev, A.V.; Dzantiev, B.B.; Eremin, S.A. Fluorescence Polarization-Based Bioassays: New Horizons. Sensors 2020, 20, 7132. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Tao, J.; Uppal, J.S.; Peng, H.; Wang, H.; Le, X.C. Nucleic acid aptamers improving fluorescence anisotropy and fluorescence polarization assays for small molecules. TrAC Trends Anal. Chem. 2019, 110, 401–409. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006; p. 954. [Google Scholar]
- Perrier, S.; Guieu, V.; Chovelon, B.; Ravelet, C.; Peyrin, E. Panoply of Fluorescence Polarization/Anisotropy Signaling Mechanisms for Functional Nucleic Acid-Based Sensing Platforms. Anal. Chem. 2018, 90, 4236–4248. [Google Scholar] [CrossRef]
- Hirata, R.; Hirakawa, K.; Shimada, N.; Watanabe, K.; Ohtsuki, T. Fluorescence lifetime probes for detection of RNA degradation. Analyst 2021, 146, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Jameson, D.M.; Ross, J.A. Fluorescence polarization/anisotropy in diagnostics and imaging. Chem. Rev. 2010, 110, 2685–2708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, S.; De Ruyck, K.; Beloglazova, N.V.; Eremin, S.A.; De Saeger, S.; Zhang, S.; Shen, J.; Wang, Z. Fluorescence polarization assays for chemical contaminants in food and environmental analyses. TrAC Trends Anal. Chem. 2019, 114, 293–313. [Google Scholar] [CrossRef]
- Xiao, X.; Zhen, S. Recent advances in fluorescence anisotropy/polarization signal amplification. RSC Adv. 2022, 12, 6364–6376. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; McConnell, E.M.; Cruz-Toledo, J.; Bernard, E.D.; Pach, A.; Mastronardi, E.; Zhang, X.; Beking, M.; Francis, T.; Giamberardino, A.; et al. Analysis of In Vitro Aptamer Selection Parameters. J. Mol. Evol. 2015, 81, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.F.; Ling, M.; Kacherovsky, N.; Pun, S.H. Aptamers 101: Aptamer discovery and in vitro applications in biosensors and separations. Chem. Sci. 2023, 14, 4961–4978. [Google Scholar] [CrossRef]
- Shaban, S.M.; Kim, D.-H. Recent Advances in Aptamer Sensors. Sensors 2021, 21, 979. [Google Scholar] [CrossRef]
- Ni, S.; Zhuo, Z.; Pan, Y.; Yu, Y.; Li, F.; Liu, J.; Wang, L.; Wu, X.; Li, D.; Wan, Y.; et al. Recent Progress in Aptamer Discoveries and Modifications for Therapeutic Applications. ACS Appl. Mater. Interfaces 2021, 13, 9500–9519. [Google Scholar] [CrossRef] [PubMed]
- Marin-Gonzalez, A.; Vilhena, J.G.; Perez, R.; Moreno-Herrero, F. A molecular view of DNA flexibility. Q. Rev. Biophys. 2021, 54, e8. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhao, Q. Direct fluorescence anisotropy approach for aflatoxin B1 detection and affinity binding study by using single tetramethylrhodamine labeled aptamer. Talanta 2018, 189, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, Q. Aptamer Structure Switch Fluorescence Anisotropy Assay for Small Molecules Using Streptavidin as an Effective Signal Amplifier Based on Proximity Effect. Anal. Chem. 2019, 91, 7379–7384. [Google Scholar] [CrossRef] [PubMed]
- Samokhvalov, A.V.; Safenkova, I.V.; Eremin, S.A.; Zherdev, A.V.; Dzantiev, B.B. Use of anchor protein modules in fluorescence polarisation aptamer assay for ochratoxin A determination. Anal. Chim. Acta 2017, 962, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Bloomfield, V.A. DNA condensation by multivalent cations. Biopolymers 1997, 44, 269–282. [Google Scholar] [CrossRef]
- Sykes, K.S.; Oliveira, L.F.L.; Stan, G.; White, R.J. Electrochemical Studies of Cation Condensation-Induced Collapse of Surface-Bound DNA. Langmuir 2019, 35, 12962–12970. [Google Scholar] [CrossRef] [PubMed]
- Froehlich, E.; Mandeville, J.S.; Arnold, D.; Kreplak, L.; Tajmir-Riahi, H.A. PEG and mPEG-anthracene induce DNA condensation and particle formation. J. Phys. Chem. B 2011, 115, 9873–9879. [Google Scholar] [CrossRef] [PubMed]
- Nikiforov, T.T.; Jeong, S. Detection of hybrid formation between peptide nucleic acids and DNA by fluorescence polarization in the presence of polylysine. Anal. Biochem. 1999, 275, 248–253. [Google Scholar] [CrossRef]
- Hayashi, K.; Chaya, H.; Fukushima, S.; Watanabe, S.; Takemoto, H.; Osada, K.; Nishiyama, N.; Miyata, K.; Kataoka, K. Influence of RNA Strand Rigidity on Polyion Complex Formation with Block Catiomers. Macromol. Rapid. Commun. 2016, 37, 486–493. [Google Scholar] [CrossRef]
- Safenkova, I.V.; Samokhvalov, A.V.; Serebrennikova, K.V.; Eremin, S.A.; Zherdev, A.V.; Dzantiev, B.B. DNA Probes for Cas12a-Based Assay with Fluorescence Anisotropy Enhanced Due to Anchors and Salts. Biosensors 2023, 13, 1034. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Meisburger, S.P.; Pabit, S.A.; Sutton, J.L.; Webb, W.W.; Pollack, L. Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc. Natl. Acad. Sci. USA 2012, 109, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Hamad, G.M.; Mehany, T.; Simal-Gandara, J.; Abou-Alella, S.; Esua, O.J.; Abdel-Wahhab, M.A.; Hafez, E.E. A review of recent innovative strategies for controlling mycotoxins in foods. Food Control 2023, 144, 109350. [Google Scholar] [CrossRef]
- Le, L.C.; Cruz-Aguado, J.A.; Penner, G.A. DNA Ligands for Aflatoxin and Zearalenone. WO2011020198A1, 24 February 2011. [Google Scholar]
- Xu, G.; Wang, C.; Yu, H.; Li, Y.; Zhao, Q.; Zhou, X.; Li, C.; Liu, M. Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer. Nucleic Acids Res. 2023, 51, 7666–7674. [Google Scholar] [CrossRef] [PubMed]
- Beloglazova, N.V.; Eremin, S.A. Rapid screening of aflatoxin B1 in beer by fluorescence polarization immunoassay. Talanta 2015, 142, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.J.; Eremin, S.; Mi, T.J.; Zhang, S.X.; Shen, J.Z.; Wang, Z.H. The development of a fluorescence polarization immunoassay for aflatoxin detection. Biomed. Environ. Sci. BES 2014, 27, 126–129. [Google Scholar] [PubMed]
- Yang, X.X.G.; Zhang, X.; Li, D.; Li, H.; Sun, Y.; Zhang, F.; Liu, C. A fluorescence polarization immunoassay for the detection of aflatoxins in herbal teas. Acta Pharm. Sin. 2017, 52, 620–624. [Google Scholar]
- Li, Y.; Yu, H.; Zhao, Q. Aptamer fluorescence anisotropy assays for detection of aflatoxin B1 and adenosine triphosphate using antibody to amplify signal change. RSC Adv. 2022, 12, 7464–7468. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, Q. Aptamer structure switch fluorescence anisotropy assay for aflatoxin B1 using tetramethylrhodamine-guanine interaction to enhance signal change. Chin. Chem. Lett. 2020, 31, 1982–1985. [Google Scholar] [CrossRef]
- Samokhvalov, A.V.; Safenkova, I.V.; Eremin, S.A.; Zherdev, A.V.; Dzantiev, B.B. Measurement of (Aptamer-Small Target) KD Using the Competition between Fluorescently Labeled and Unlabeled Targets and the Detection of Fluorescence Anisotropy. Anal. Chem. 2018, 90, 9189–9198. [Google Scholar] [CrossRef]
- Sebaugh, J.L.; McCray, P.D. Defining the linear portion of a sigmoid—Shaped curve: Bend points. Pharm. Stat. 2003, 2, 167–174. [Google Scholar] [CrossRef]
- Wang, D.; Zhu, J.; Zhang, Z.; Zhang, Q.; Zhang, W.; Yu, L.; Jiang, J.; Chen, X.; Wang, X.; Li, P. Simultaneous Lateral Flow Immunoassay for Multi-Class Chemical Contaminants in Maize and Peanut with One-Stop Sample Preparation. Toxins 2019, 11, 56. [Google Scholar] [CrossRef] [PubMed]
- Romero-Sanchez, I.; Ramirez-Garcia, L.; Gracia-Lor, E.; Madrid-Albarran, Y. Simultaneous determination of aflatoxins B1, B2, G1 and G2 in commercial rices using immunoaffinity column clean-up and HPLC-MS/MS. Food Chem. 2022, 395, 133611. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Kirihata, T.; Fujii, S.; Sakai, H.; Kuwahara, M.; Sawai, H.; Sugimoto, N. Influence of cationic molecules on the hairpin to duplex equilibria of self-complementary DNA and RNA oligonucleotides. Nucleic Acids Res. 2007, 35, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, L.; Zhao, Q. Development of aptamer fluorescent switch assay for aflatoxin B1 by using fluorescein-labeled aptamer and black hole quencher 1-labeled complementary DNA. Anal. Bioanal. Chem. 2018, 410, 6269–6277. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Lee, K.; Li, S. Nucleic Acids Based Polyelectrolyte Complexes: Their Complexation Mechanism, Morphology, and Stability. Chem. Mater. 2021, 33, 7923–7943. [Google Scholar] [CrossRef]
- Cheng, C.; Jia, J.L.; Ran, S.Y. Polyethylene glycol and divalent salt-induced DNA reentrant condensation revealed by single molecule measurements. Soft Matter 2015, 11, 3927–3935. [Google Scholar] [CrossRef] [PubMed]
- Katayose, S.; Kataoka, K. Water-soluble polyion complex associates of DNA and poly(ethylene glycol)-poly(L-lysine) block copolymer. Bioconjugate Chem. 1997, 8, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Bonner, G.; Klibanov, A.M. Structural stability of DNA in nonaqueous solvents. Biotechnol. Bioeng. 2000, 68, 339–344. [Google Scholar] [CrossRef]
- Amirbekyan, K.Y.; Shahinyan, G.A.; Ghazoyan, H.H.; Sargsyan, H.R.; Markarian, S.A. Fluorescence anisotropy studies on the Hoechst 33258-DNA interaction: The solvent effect. J. Biomol. Struct. Dyn. 2021, 39, 4902–4906. [Google Scholar] [CrossRef]
- Xu, M.; Dai, T.; Wang, Y.; Yang, G. The incipient denaturation mechanism of DNA. RSC Adv. 2022, 12, 23356–23365. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.D.; Adami, R.C. Stabilization of DNA utilizing divalent cations and alcohol. Int. J. Pharm. 2003, 264, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Mehta, S.K. Developments of Polysorbate (Tween) based microemulsions: Preclinical drug delivery, toxicity and antimicrobial applications. Int. J. Pharm. 2017, 529, 134–160. [Google Scholar] [CrossRef]
- Commission Regulation (EU) 2023/915 of 25 April 2023 on Maximum Levels for Certain Contaminants in Food and Repealing Regulation (EC) No 1881/2006. Available online: https://eur-lex.europa.eu/eli/reg/2023/915/oj (accessed on 20 March 2024).
- Vita, V.; Franchino, C.; Iammarino, M.; De Pace, R. Aflatoxins contamination in nuts for direct human consumption: Analytical findings from three years of official control in Italy. Int. J. Food Sci. Technol. 2022, 57, 7496–7504. [Google Scholar] [CrossRef]
- Tawani, A.; Mishra, S.K.; Kumar, A. Structural insight for the recognition of G-quadruplex structure at human c-myc promoter sequence by flavonoid Quercetin. Sci. Rep. 2017, 7, 3600. [Google Scholar] [CrossRef]
- Ye, H.; Lu, Q.; Duan, N.; Wang, Z. GO-amplified fluorescence polarization assay for high-sensitivity detection of aflatoxin B1 with low dosage aptamer probe. Anal. Bioanal. Chem. 2019, 411, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Qin, J.; Hu, K.; Liu, X.; Zhao, S.; Huang, Y. Novel autonomous protein-encoded aptamer nanomachines and isothermal exponential amplification for ultrasensitive fluorescence polarization sensing of small molecules. RSC Adv. 2016, 6, 86043–86050. [Google Scholar] [CrossRef]
- Nasir, M.S.; Jolley, M.E. Development of a fluorescence polarization assay for the determination of aflatoxins in grains. J. Agric. Food Chem. 2002, 50, 3116–3121. [Google Scholar] [CrossRef]
- Raysyan, A.; Eremin, S.A.; Beloglazova, N.V.; De Saeger, S.; Gravel, I.V. Immunochemical approaches for detection of aflatoxin B1 in herbal medicines. Phytochem. Anal. 2020, 31, 662–669. [Google Scholar] [CrossRef]
- Zezza, F.; Longobardi, F.; Pascale, M.; Eremin, S.A.; Visconti, A. Fluorescence polarization immunoassay for rapid screening of ochratoxin A in red wine. Anal. Bioanal. Chem. 2009, 395, 1317–1323. [Google Scholar] [CrossRef]
- Zhao, Z.; Wei, L.; Cao, M.; Lu, M. A smartphone-based system for fluorescence polarization assays. Biosens. Bioelectron. 2019, 128, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Aoyagi, M.; Fukuyama, M.; Maeki, M.; Ishida, A.; Tani, H.; Shigemura, K.; Hibara, A.; Tokeshi, M. Determination of Deoxynivalenol in Wheat, Barley, Corn Meal, and Wheat-Based Products by Simultaneous Multisample Fluorescence Polarization Immunoassay Using a Portable Analyzer. ACS Food Sci. Technol. 2021, 1, 1623–1628. [Google Scholar] [CrossRef]






| Lines | Short Indications | Oligonucleotide Sequences | Stem Length | *** H-Bonds | KD (nM) |
|---|---|---|---|---|---|
| 1 | Initial (untruncated) | GTT GGG CAC GTG TTG TCT CTC TGT GTC T * CG TGC CCT TCG CTA GGC CCA CA ** | 7 | 20 | 173.1 ± 9.5 |
| 2 | 22 nt | C GTG TTG TCT CTC TGT GTC TCG | 2 | 6 | 2751 ± 443 **** |
| 3 | 26 nt | CAC GTG TTG TCT CTC TGT GTC TCG TG | 4 | 11 | 271.8 ± 51.1 **** |
| 4 | 32 nt | GGG CAC GTG TTG TCT CTC TGT GTC TCG TGC CC | 7 | 20 | 163.7 ± 19.0 |
| 5 | 38 nt | GTT GGG CAC GTG TTG TCT CTC TGT GTC TCG TGC CCA AC | 10 | 27 | 123.0 ± 24.4 |
| 6 | 42 nt | GG GTT GGG CAC GTTG TTG TCT CTC TGT GTC TCG TGC CCA ACC C | 12 | 33 | 218.0 ± 13.2 |
| 7 | 46 nt | T TGG GTT GGG CAC GTG TTG TCT CTC TGT GTC TCG TGC CCA ACC CAA | 14 | 37 | 147.5 ± 14.5 |
| 8 | 54 nt | T TGG TTG G GTT GGG CAC GTG TTG TCT CTC TGT GTC TCG TGC CCA ACC CAA CCA A | 18 | 47 | 559.7 ± 45.3 |
| Sample | Added (μg/kg) | Found (μg/kg) | Recovery (%) |
|---|---|---|---|
| Almond flour | 200 | 242.1 ± 50.5 | 121.0 |
| 100 | 121.2 ± 33.5 | 121.1 | |
| 75 | 91.6 ± 20.1 | 122.1 | |
| 30 | 33.8 ± 15.8 | 112.7 | |
| 10 | 12.3 ± 4.6 | 123.3 |
| Sample | Found AFB1 (μg/kg), HPLC-MS/MS Technique | Found AFB1 (μg/kg), FA-Based Assay | Recovery for FA-Based Assay (%) |
|---|---|---|---|
| 1 | 376.3 ±12.6 | 416.1 ± 24.9 | 110.6 ± 6.0 |
| 2 | 282.2 ± 9.6 | 302.2 ± 9.1 | 107.1 ± 3.0 |
| 3 | 188.2 ± 4.5 | 212.9 ± 26.7 | 113.2 ± 12,5 |
| 4 | 112.9 ± 0.7 | 118.6 ± 19.3 | 105.0 ± 16.3 |
| 5 | 89.4 ± 0.3 | 75.0 ± 10.2 | 83.9 ± 13.7 |
| 6 | 37.8 ± 2.2 | 39.7 ± 10.4 | 105.1 ± 26.2 |
| 7 | not detected | <LOD | - |
| 8 | not detected | <LOD | - |
| Methods | Complexity | Linear (L)/Dynamic (D) Range (ng/mL) | LOD (ng/mL) | Tested Samples | Reference |
|---|---|---|---|---|---|
| Immunoassays | |||||
| Competition between native ligand and labeled ligand | One-step | - | 5 | Popcorn, corn, sorghum, peanut paste, peanut butter | [51] |
| One-step | (L) 16.25–33.49 | 13.12 | - | [28] | |
| One-step | (L) 3–84 | 1 | beer | [27] | |
| One-step | (L) 92.76–252.32 | 20 | tea leaves | [29] | |
| One-step | (L) 8.6–63.7 | 1 | leaves of medicinal plants | [52] | |
| Aptamer assays | |||||
| Polycation-assisted competitive aptamer assay with labeled ligand | One-step | (L) 4.0 ± 0.4–137.8 ± 8.0 | 0.72 ± 0.3 | Almond flour | This work |
| Competitive aptamer assay with labeled ligand | One-step | (L) 15.0 ± 3.4–117.4 ± 11.2 | 9.7 ± 1.6 | Almond flour | This work |
| Protein-encoded aptamer nanomachines and isothermal exponential amplification | Multi-step | - | 0.000078 | - | [50] |
| Direct labeling with guanine sensitive tetramethylrhodamine label | One-step | (D) 0.62–312.27 | 0.62 | Serum; urine, wine, beer | [14] |
| Ligand-induced strand displacement with streptavidin anchor | Multi-step | (D) 0.08–39 | 0.019 | White | [15] |
| Physical absorption of fluorescein-labeled aptamer on graphene oxide | Multi-step | (L) 0.015–1.56 | 0.015 | Rice | [49] |
| Direct labeling with guanine sensitive tetramethylrhodamine label enhanced by strand displacement of guanine-rich ssDNA | One-step | (D) 0.04–9.74 | 0.039 | Grape juice, milk, tap water | [31] |
| Ligand-induced strand displacement with IgG anchor | Multi-step | (D) 0.008–31.2 | 0.008 | Grape juice, white wine, tap water | [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samokhvalov, A.V.; Mironova, A.A.; Eremin, S.A.; Zherdev, A.V.; Dzantiev, B.B. Polycations as Aptamer-Binding Modulators for Sensitive Fluorescence Anisotropy Assay of Aflatoxin B1. Sensors 2024, 24, 3230. https://doi.org/10.3390/s24103230
Samokhvalov AV, Mironova AA, Eremin SA, Zherdev AV, Dzantiev BB. Polycations as Aptamer-Binding Modulators for Sensitive Fluorescence Anisotropy Assay of Aflatoxin B1. Sensors. 2024; 24(10):3230. https://doi.org/10.3390/s24103230
Chicago/Turabian StyleSamokhvalov, Alexey V., Alena A. Mironova, Sergei A. Eremin, Anatoly V. Zherdev, and Boris B. Dzantiev. 2024. "Polycations as Aptamer-Binding Modulators for Sensitive Fluorescence Anisotropy Assay of Aflatoxin B1" Sensors 24, no. 10: 3230. https://doi.org/10.3390/s24103230







