Comprehensive Pan-Cancer Analysis of Connexin 43 as a Potential Biomarker and Therapeutic Target in Human Kidney Renal Clear Cell Carcinoma (KIRC)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Analysis of Cx43 Expression in Pan-Cancer
2.2. Diagnostic Value Analysis
2.3. Survival Prognosis Analysis
2.4. Associations between GJA1 Expression and Different Clinical Features in KIRC
2.5. Univariate and Multivariate Cox Regression Analyses in KIRC
2.6. Cell Lines and Cell Culture
2.7. The mRNA Expressions of GJA1
2.8. Wound Healing Assay
2.9. DEGs between High and Low GJA1 Expression Groups in KIRC
3. Results
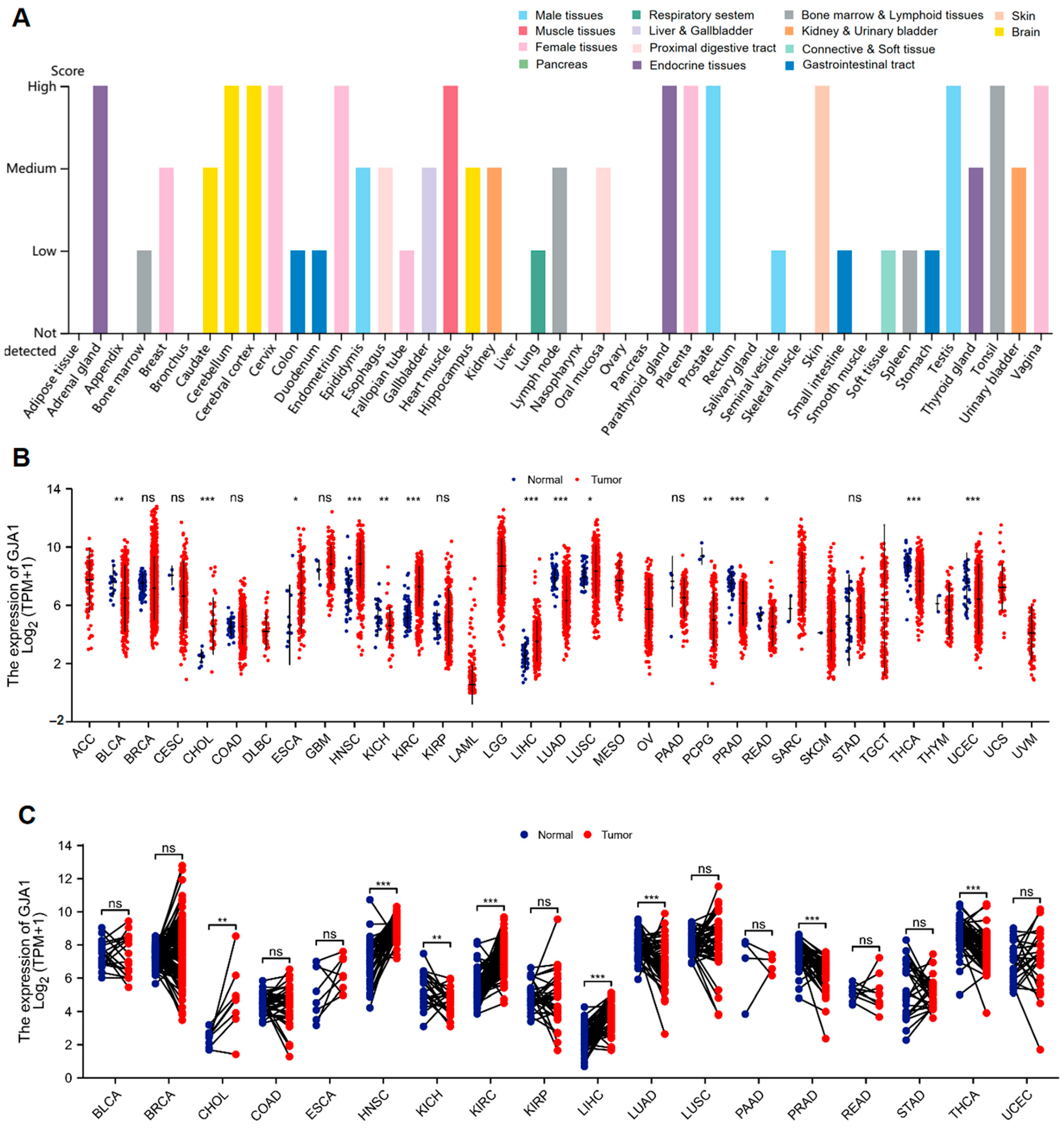
3.1. GJA1 Expression between Normal and Tumor Tissues
3.2. Diagnostic Significance of GJA1 in Cancer
3.3. Prognostic Significance of GJA1 in Cancer
3.4. GJA1 Expression Is Associated with Various Clinical Features in KIRC
3.5. Univariate and Multivariate Cox Regression Analyses in KIRC
3.6. The Migratory Capacity of KIRC Cancer Cells Were Inhibited When the Expression of GJA1 Was Elevated
3.7. DEGs between GJA1 High Expression Group and Low Expression Groups in KIRC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Valiunas, V.; Polosina, Y.Y.; Miller, H.; Potapova, I.A.; Valiuniene, L.; Doronin, S.; Mathias, R.T.; Robinson, R.B.; Rosen, M.R.; Cohen, I.S.; et al. Connexin-specific cell-to-cell transfer of short interfering RNA by gap junctions. J. Physiol. 2005, 568, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Totland, M.Z.; Rasmussen, N.L.; Knudsen, L.M.; Leithe, E. Regulation of gap junction intercellular communication by connexin ubiquitination: Physiological and pathophysiological implications. Cell Mol. Life Sci. 2020, 77, 573–591. [Google Scholar] [CrossRef]
- Danesh-Meyer, H.V.; Green, C.R. Focus on molecules: Connexin 43—Mind the gap. Exp. Eye Res. 2008, 87, 494–495. [Google Scholar] [CrossRef]
- Laird, D.W. Life cycle of connexins in health and disease. Biochem. J. 2006, 394, 527–543. [Google Scholar] [CrossRef]
- Aasen, T.; Mesnil, M.; Naus, C.C.; Lampe, P.D.; Laird, D.W. Gap junctions and cancer: Communicating for 50 years. Nat. Rev. Cancer 2016, 16, 775–788. [Google Scholar] [CrossRef]
- Zhang, J.; Riquelme, M.A.; Hua, R.; Acosta, F.M.; Gu, S.; Jiang, J.X. Connexin 43 hemichannels regulate mitochondrial ATP generation, mobilization, and mitochondrial homeostasis against oxidative stress. eLife 2022, 11, e82206. [Google Scholar] [CrossRef]
- Martins-Marques, T.; Ribeiro-Rodrigues, T.; Batista-Almeida, D.; Aasen, T.; Kwak, B.R.; Girao, H. Biological Functions of Connexin43 Beyond Intercellular Communication. Trends Cell Biol. 2019, 29, 835–847. [Google Scholar] [CrossRef]
- Boengler, K.; Schulz, R.; Heusch, G. Connexin 43 signalling and cardioprotection. Heart 2006, 92, 1724–1727. [Google Scholar] [CrossRef]
- Bonacquisti, E.E.; Nguyen, J. Connexin 43 (Cx43) in cancer: Implications for therapeutic approaches via gap junctions. Cancer Lett. 2019, 442, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Leithe, E.; Mesnil, M.; Aasen, T. The connexin 43 C-terminus: A tail of many tales. Biochim. Biophys. Acta Biomembr. 2018, 1860, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Esseltine, J.L.; Laird, D.W. Next-Generation Connexin and Pannexin Cell Biology. Trends Cell Biol. 2016, 26, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Buratto, D.; Donati, V.; Zonta, F.; Mammano, F. Harnessing the therapeutic potential of antibodies targeting connexin hemichannels. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166047. [Google Scholar] [CrossRef] [PubMed]
- Michela, P.; Velia, V.; Aldo, P.; Ada, P. Role of connexin 43 in cardiovascular diseases. Eur. J. Pharmacol. 2015, 768, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Bodenstine, T.M. Connexins and Glucose Metabolism in Cancer. Int. J. Mol. Sci. 2022, 23, 10172. [Google Scholar] [CrossRef] [PubMed]
- Alaga, K.C.; Crawford, M.; Dagnino, L.; Laird, D.W. Aberrant Cx43 Expression and Mislocalization in Metastatic Human Melanomas. J. Cancer 2017, 8, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.L.; Wang, B.R.; Chen, G.Q.; Li, G.H.; Xu, Y.Y. Clinical significance of vascular endothelial growth factor and connexin43 for predicting pancreatic cancer clinicopathologic parameters. Med. Oncol. 2010, 27, 1164–1170. [Google Scholar] [CrossRef] [PubMed]
- Georgikou, C.; Yin, L.; Gladkich, J.; Xiao, X.; Sticht, C.; Torre, C.; Gretz, N.; Gross, W.; Schäfer, M.; Karakhanova, S.; et al. Inhibition of miR30a-3p by sulforaphane enhances gap junction intercellular communication in pancreatic cancer. Cancer Lett. 2020, 469, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, H.; Wang, E. Expressions of connexin 43 and E-cadherin and their correlation in non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2005, 8, 103–106. [Google Scholar] [PubMed]
- Huang, W.; Wang, Y.; He, T.; Zhu, J.; Li, J.; Zhang, S.; Zhu, Y.; Xu, Y.; Xu, L.; Wang, H.; et al. Arteannuin B Enhances the Effectiveness of Cisplatin in Non-Small Cell Lung Cancer by Regulating Connexin 43 and MAPK Pathway. Am. J. Chin. Med. 2022, 50, 1963–1992. [Google Scholar] [CrossRef]
- Nesmiyanov, P.P.; Tolkachev, B.E.; Strygin, A.V. ZO-1 expression shows prognostic value in chronic B cell leukemia. Immunobiology 2016, 221, 6–11. [Google Scholar] [CrossRef]
- Lamiche, C.; Clarhaut, J.; Strale, P.O.; Crespin, S.; Pedretti, N.; Bernard, F.X.; Naus, C.C.; Chen, V.C.; Foster, L.J.; Defamie, N.; et al. The gap junction protein Cx43 is involved in the bone-targeted metastatic behaviour of human prostate cancer cells. Clin. Exp. Metastasis 2012, 29, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Kazan, J.M.; El-Saghir, J.; Saliba, J.; Shaito, A.; Jalaleddine, N.; El-Hajjar, L.; Al-Ghadban, S.; Yehia, L.; Zibara, K.; El-Sabban, M. Cx43 Expression Correlates with Breast Cancer Metastasis in MDA-MB-231 Cells In Vitro, In a Mouse Xenograft Model and in Human Breast Cancer Tissues. Cancers 2019, 11, 460. [Google Scholar] [CrossRef] [PubMed]
- Sinha, G.; Ferrer, A.I.; Moore, C.A.; Naaldijk, Y.; Rameshwar, P. Gap Junctions and Breast Cancer Dormancy. Trends Cancer 2020, 6, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.L.; Williams, C.B.; Zambrano, J.N.; Williams, C.J.; Yeh, E.S. Connexin 43 in the development and progression of breast cancer: What’s the connection? (Review). Int. J. Oncol. 2017, 51, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Chasampalioti, M.; Green, A.R.; Ellis, I.O.; Rakha, E.A.; Jackson, A.M.; Spendlove, I.; Ramage, J.M. Connexin 43 is an independent predictor of patient outcome in breast cancer patients. Breast Cancer Res. Treat. 2019, 174, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Zefferino, R.; Piccoli, C.; Gioia, S.D.; Capitanio, N.; Conese, M. Gap Junction Intercellular Communication in the Carcinogenesis Hallmarks: Is This a Phenomenon or Epiphenomenon? Cells 2019, 8, 896. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.I.; Wang, L.H. Emerging roles of gap junction proteins connexins in cancer metastasis, chemoresistance and clinical application. J. Biomed. Sci. 2019, 26, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.Z.; Riquelme, M.A.; Gu, S.; Kar, R.; Gao, X.; Sun, L.; Jiang, J.X. Osteocytic connexin hemichannels suppress breast cancer growth and bone metastasis. Oncogene 2016, 35, 5597–5607. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Z.Y.; Guo, Y.F.; Kuang, J.Y.; Bian, X.W.; Yu, S.C. Targeting different domains of gap junction protein to control malignant glioma. Neuro Oncol. 2018, 20, 885–896. [Google Scholar] [CrossRef]
- Yang, Z.J.; Bi, Q.C.; Gan, L.J.; Zhang, L.L.; Wei, M.J.; Hong, T.; Liu, R.; Qiu, C.L.; Han, X.J.; Jiang, L.P. Exosomes Derived from Glioma Cells under Hypoxia Promote Angiogenesis through Up-regulated Exosomal Connexin 43. Int. J. Med. Sci. 2022, 19, 1205–1215. [Google Scholar] [CrossRef]
- Kanczuga-Koda, L.; Sulkowski, S.; Lenczewski, A.; Koda, M.; Wincewicz, A.; Baltaziak, M.; Sulkowska, M. Increased expression of connexins 26 and 43 in lymph node metastases of breast cancer. J. Clin. Pathol. 2006, 59, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.A.; Ilina, O.; Vasaturo, A.; Venhuizen, J.H.; Vullings, M.; Venhuizen, V.; Bilos, A.; Figdor, C.G.; Span, P.N.; Friedl, P. Collective invasion induced by an autocrine purinergic loop through connexin-43 hemichannels. J. Cell Biol. 2020, 219, e201911120. [Google Scholar] [CrossRef] [PubMed]
- Stoletov, K.; Strnadel, J.; Zardouzian, E.; Momiyama, M.; Park, F.D.; Kelber, J.A.; Pizzo, D.P.; Hoffman, R.; VandenBerg, S.R.; Klemke, R.L. Role of connexins in metastatic breast cancer and melanoma brain colonization. J. Cell Sci. 2013, 126, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Grek, C.L.; Rhett, J.M.; Bruce, J.S.; Ghatnekar, G.S.; Yeh, E.S. Connexin 43, breast cancer tumor suppressor: Missed connections? Cancer Lett. 2016, 374, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.F.; Varghese, R.T.; Lamouille, S.; Guo, S.; Pridham, K.J.; Kanabur, P.; Osimani, A.M.; Sharma, S.; Jourdan, J.; Rodgers, C.M.; et al. Connexin 43 Inhibition Sensitizes Chemoresistant Glioblastoma Cells to Temozolomide. Cancer Res. 2016, 76, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Sirnes, S.; Bruun, J.; Kolberg, M.; Kjenseth, A.; Lind, G.E.; Svindland, A.; Brech, A.; Nesbakken, A.; Lothe, R.A.; Leithe, E.; et al. Connexin43 acts as a colorectal cancer tumor suppressor and predicts disease outcome. Int. J. Cancer 2012, 131, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Leithe, E.; Graham, S.V.; Kameritsch, P.; Mayán, M.D.; Mesnil, M.; Pogoda, K.; Tabernero, A. Connexins in cancer: Bridging the gap to the clinic. Oncogene 2019, 38, 4429–4451. [Google Scholar] [CrossRef]
- Wang, Z.S.; Wu, L.Q.; Yi, X.; Geng, C.; Li, Y.J.; Yao, R.Y. Connexin-43 can delay early recurrence and metastasis in patients with hepatitis B-related hepatocellular carcinoma and low serum alpha-fetoprotein after radical hepatectomy. BMC Cancer 2013, 13, 306. [Google Scholar] [CrossRef]
- Xu, N.; Chen, H.J.; Chen, S.H.; Xue, X.Y.; Chen, H.; Zheng, Q.S.; Wei, Y.; Li, X.D.; Huang, J.B.; Cai, H.; et al. Reduced Connexin 43 expression is associated with tumor malignant behaviors and biochemical recurrence-free survival of prostate cancer. Oncotarget 2016, 7, 67476–67484. [Google Scholar] [CrossRef]
- Cliff, C.L.; Williams, B.M.; Chadjichristos, C.E.; Mouritzen, U.; Squires, P.E.; Hills, C.E. Connexin 43: A Target for the Treatment of Inflammation in Secondary Complications of the Kidney and Eye in Diabetes. Int. J. Mol. Sci. 2022, 23, 600. [Google Scholar] [CrossRef]
- Zhang, W.; Lin, L.; Zhang, Y.; Zhao, T.; Zhan, Y.; Wang, H.; Fang, J.; Du, B. Dioscin potentiates the antitumor effect of suicide gene therapy in melanoma by gap junction intercellular communication-mediated antigen cross-presentation. Biomed. Pharmacother. 2022, 150, 112973. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, M.; Li, Y.; Shi, M.; Wang, Z.; Cao, C.; Hong, Y.; Hu, B.; Zhu, H.; Zhao, Z.; et al. Blocking connexin 43 and its promotion of ATP release from renal tubular epithelial cells ameliorates renal fibrosis. Cell Death Dis. 2022, 13, 511. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef] [PubMed]
- Vanderpuye, O.A.; Bell, C.L.; Murray, S.A. Redistribution of connexin 43 during cell division. Cell Biol. Int. 2016, 40, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T. Connexins: Junctional and non-junctional modulators of proliferation. Cell Tissue Res. 2015, 360, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Gleisner, M.A.; Navarrete, M.; Hofmann, F.; Salazar-Onfray, F.; Tittarelli, A. Mind the Gaps in Tumor Immunity: Impact of Connexin-Mediated Intercellular Connections. Front. Immunol. 2017, 8, 1067. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, H.; Sato, H.; Shirai, S.; Kobayashi, S.; Fukumoto, K.; Ishida, T.; Seki, T.; Ariga, T.; Yano, T. Connexin 32 down-regulates the fibrinolytic factors in metastatic renal cell carcinoma cells. Life Sci. 2006, 78, 2249–2254. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Sekine, M.; Kobayashi, M.; Virgona, N.; Ota, M.; Yano, T. Induction of the connexin 32 gene by epigallocatechin-3-gallate potentiates vinblastine-induced cytotoxicity in human renal carcinoma cells. Chemotherapy 2013, 59, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, E.; Sato, H.; Nagashima, Y.; Negishi, E.; Shirai, S.; Fukumoto, K.; Hagiwara, H.; Hagiwara, K.; Ueno, K.; Yano, T. A Src family inhibitor (PP1) potentiates tumor-suppressive effect of connexin 32 gene in renal cancer cells. Life Sci. 2005, 76, 2711–2720. [Google Scholar] [CrossRef]
- Chen, Q.; Boire, A.; Jin, X.; Valiente, M.; Er, E.E.; Lopez-Soto, A.; Jacob, L.; Patwa, R.; Shah, H.; Xu, K.; et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 2016, 533, 493–498. [Google Scholar] [CrossRef]
- Munoz, J.L.; Rodriguez-Cruz, V.; Greco, S.J.; Ramkissoon, S.H.; Ligon, K.L.; Rameshwar, P. Temozolomide resistance in glioblastoma cells occurs partly through epidermal growth factor receptor-mediated induction of connexin 43. Cell Death Dis. 2014, 5, e1145. [Google Scholar] [CrossRef]
- Osswald, M.; Jung, E.; Sahm, F.; Solecki, G.; Venkataramani, V.; Blaes, J.; Weil, S.; Horstmann, H.; Wiestler, B.; Syed, M.; et al. Brain tumour cells interconnect to a functional and resistant network. Nature 2015, 528, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, J.; Nomata, K.; Noguchi, M.; Satoh, H.; Kanda, S.; Kanetake, H.; Saito, Y. All-trans retinoic acid enhances gap junctional intercellular communication among renal epithelial cells in vitro treated with renal carcinogens. Eur. J. Cancer 1999, 35, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, T.; Suzuki, Y.; Okamoto, T.; Mastushita, N.; Hasegawa, M.; Omori, S. Prevention of renal cell carcinoma by active vitamin D3. World J. Surg. 2000, 24, 1205–1210. [Google Scholar] [CrossRef]
- Liu, K.; Lai, M.; Wang, S.; Zheng, K.; Xie, S.; Wang, X. Construction of a CXC Chemokine-Based Prediction Model for the Prognosis of Colon Cancer. Biomed Res. Int. 2020, 2020, 6107865. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jiao, N.; Sun, T.; Ma, Y.; Zhang, X.; Chen, H.; Hong, J.; Zhang, Y. CXCL11 Correlates With Antitumor Immunity and an Improved Prognosis in Colon Cancer. Front. Cell Dev. Biol. 2021, 9, 646252. [Google Scholar] [CrossRef]
- Evans, W.H.; De Vuyst, E.; Leybaert, L. The gap junction cellular internet: Connexin hemichannels enter the signalling limelight. Biochem. J. 2006, 397, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, H.; Chen, H.; Tan, T.; Wang, Y.; Yang, H.; Ding, Y.; Wang, S. CX43 down-regulation promotes cell aggressiveness and 5-fluorouracil-resistance by attenuating cell stiffness in colorectal carcinoma. Cancer Biol. Ther. 2023, 24, 2221879. [Google Scholar] [CrossRef]
- Corso, G.; Figueiredo, J.; De Angelis, S.P.; Corso, F.; Girardi, A.; Pereira, J.; Seruca, R.; Bonanni, B.; Carneiro, P.; Pravettoni, G.; et al. E-cadherin deregulation in breast cancer. J. Cell. Mol. Med. 2020, 24, 5930–5936. [Google Scholar] [CrossRef] [PubMed]
- Shu, G.; Lu, X.; Pan, Y.; Cen, J.; Huang, K.; Zhou, M.; Lu, J.; Dong, J.; Han, H.; Chen, W.; et al. Exosomal circSPIRE1 mediates glycosylation of E-cadherin to suppress metastasis of renal cell carcinoma. Oncogene 2023, 42, 1802–1820. [Google Scholar] [CrossRef]
- Wen, J.; Min, X.; Shen, M.; Hua, Q.; Han, Y.; Zhao, L.; Liu, L.; Huang, G.; Liu, J.; Zhao, X. ACLY facilitates colon cancer cell metastasis by CTNNB1. J. Exp. Clin. Cancer Res. 2019, 38, 401. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Fan, M.Q.; Xie, X.X.; Shu, Q.P.; Du, X.H.; Qi, L.Z.; Zhang, X.D.; Zhang, M.H.; Shan, G.; Du, R.L.; et al. Activation of CTNNB1 by deubiquitinase UCHL3-mediated stabilization facilitates bladder cancer progression. J. Transl. Med. 2023, 21, 656. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Brendel, M.; Getseva, V.; Assaad, M.A.; Sigouros, M.; Sigaras, A.; Kane, T.; Khosravi, P.; Mosquera, J.M.; Elemento, O.; Hajirasouliha, I. Weakly-supervised tumor purity prediction from frozen H&E stained slides. EBioMedicine 2022, 80, 104067. [Google Scholar]





| Characteristic | Low Expression of GJA1 | High Expression of GJA1 | p | |
|---|---|---|---|---|
| n | 269 | 270 | ||
| T stage, n (%) | 0.023 b | |||
| T1 | 121 (22.4%) | 157 (29.1%) | ||
| T2 | 39 (7.2%) | 32 (5.9%) | ||
| T3 | 103 (19.1%) | 76 (14.1%) | ||
| T4 | 6 (1.1%) | 5 (0.9%) | ||
| N stage, n (%) | 0.251 b | |||
| N0 | 122 (47.5%) | 119 (46.3%) | ||
| N1 | 11 (4.3%) | 5 (1.9%) | ||
| M stage, n (%) | 0.005 b | |||
| M0 | 202 (39.9%) | 226 (44.7%) | ||
| M1 | 51 (10.1%) | 27 (5.3%) | ||
| Age, mean ± SD | 61.62 ± 11.69 | 59.64 ± 12.43 | 0.058 a |
| Characteristics | Total (n) | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| Hazard Ratio (95% CI) | p Value | Hazard Ratio (95% CI) | p Value | ||
| Age (≤60 vs. >60) | 539 | 1.765 (1.298–2.398) | <0.001 | 1.590 (1.168–2.166) | 0.003 |
| Gender (Male vs. Female) | 539 | 1.075 (0.788–1.465) | 0.648 | ||
| Pathologic stage (Stage I and Stage II vs. Stage III and Stage IV) | 536 | 3.946 (2.872–5.423) | <0.001 | 3.654 (2.656–5.025) | <0.001 |
| Histologic grade (G1 and G2 vs. G3 and G4) | 531 | 2.702 (1.918–3.807) | <0.001 | 1.731 (1.209–2.477) | 0.003 |
| T stage (T1 and T2 vs. T3 and T4) | 539 | 3.228 (2.382–4.374) | <0.001 | 0.595 (0.331–1.070) | 0.083 |
| Laterality (left vs. right) | 538 | 0.706 (0.523–0.952) | 0.023 | 0.703 (0.519–0.951) | 0.022 |
| Treatment history (no vs. yes) | 532 | 4.401 (3.226–6.002) | <0.001 | 1.223 (0.066–22.723) | 0.893 |
| Genetic background (no vs. yes) | 533 | 0.156 (0.049–0.495) | 0.002 | 0.094 (0.010–0.848) | 0.035 |
| GJA1 (low vs. high) | 539 | 0.805 (0.719–0.901) | <0.001 | 0.758 (0.656–0.976) | 0.030 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Wang, X.; Zhu, F.; Guo, S.; Chao, Z.; Cao, C.; Lu, Z.; Zhu, H.; Wang, M.; Zhu, F.; et al. Comprehensive Pan-Cancer Analysis of Connexin 43 as a Potential Biomarker and Therapeutic Target in Human Kidney Renal Clear Cell Carcinoma (KIRC). Medicina 2024, 60, 780. https://doi.org/10.3390/medicina60050780
Xu H, Wang X, Zhu F, Guo S, Chao Z, Cao C, Lu Z, Zhu H, Wang M, Zhu F, et al. Comprehensive Pan-Cancer Analysis of Connexin 43 as a Potential Biomarker and Therapeutic Target in Human Kidney Renal Clear Cell Carcinoma (KIRC). Medicina. 2024; 60(5):780. https://doi.org/10.3390/medicina60050780
Chicago/Turabian StyleXu, Huzi, Xiuru Wang, Fan Zhu, Shuiming Guo, Zheng Chao, Chujin Cao, Zhihui Lu, Han Zhu, Meng Wang, Fengming Zhu, and et al. 2024. "Comprehensive Pan-Cancer Analysis of Connexin 43 as a Potential Biomarker and Therapeutic Target in Human Kidney Renal Clear Cell Carcinoma (KIRC)" Medicina 60, no. 5: 780. https://doi.org/10.3390/medicina60050780





