Regions of Bovine Adenovirus-3 Protein VII Involved in Interactions with Viral and Cellular Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Viruses
2.2. Antibodies
2.3. Plasmid Construction
2.4. Transfection
2.5. Western Blotting
2.6. Indirect Immunofluorescence Assay
2.7. GST Pull-Down Assay
2.8. Bimolecular Fluorescence Complementation (BiFC) Analysis
2.9. Isolation of Recombinant BAdV-3 (BAV.VII.ST)
2.10. Virus Growth
2.11. Purification and Identification of the Strep-Tagged Protein Complex
2.12. Co-Immunoprecipitation (Co-IP) Assay
3. Results
3.1. Sequence Analysis of BAdV-3 Protein VII
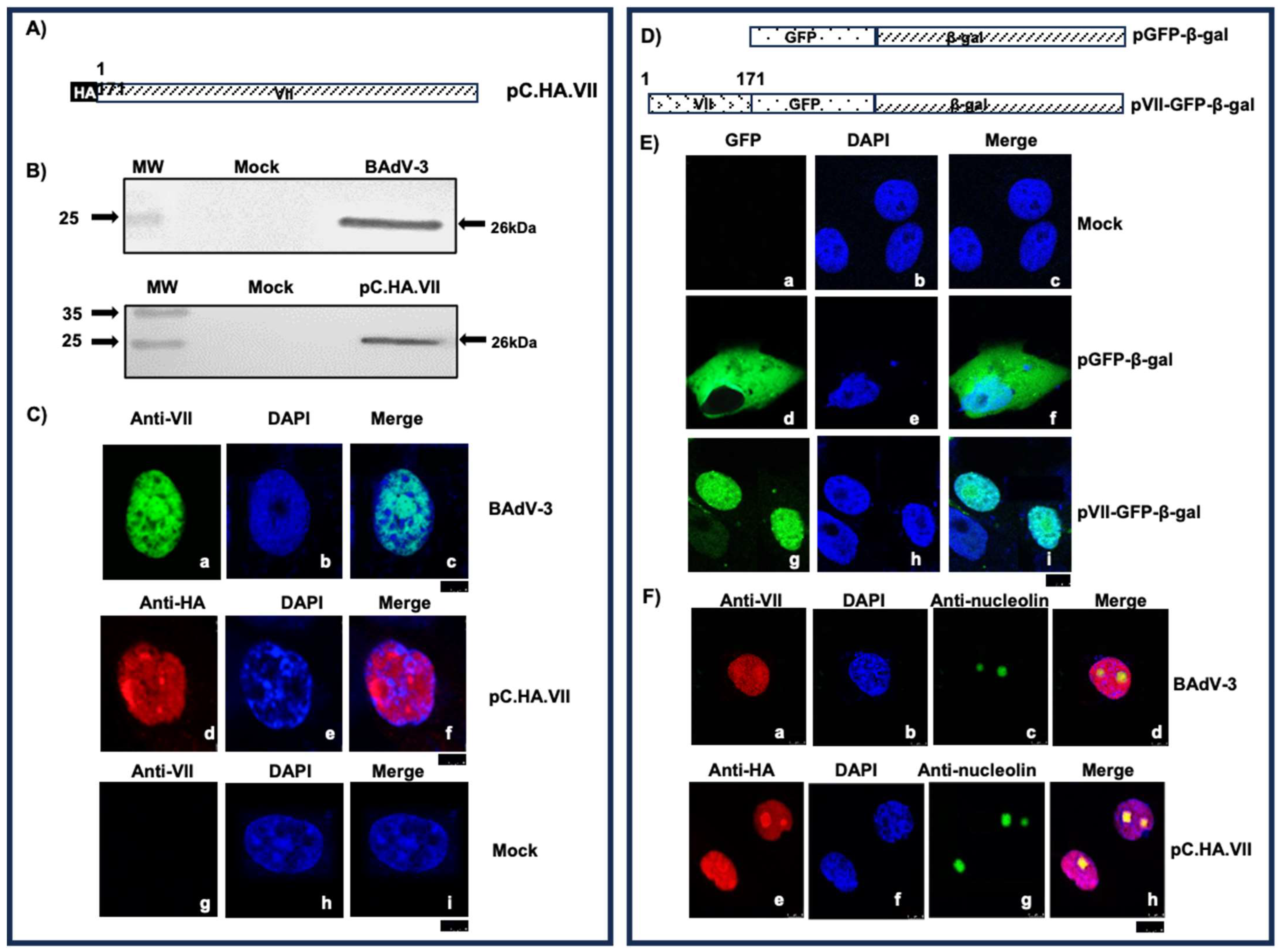
3.2. Characterization of Protein VII
3.3. Identification of Functional Nuclear/Nucleolar Localization Sequences of Protein VII
3.4. Interaction of Protein VII with Importins/Transportin
3.5. Construction of BAdV-3 with Strep-Tagged Protein VII
3.6. Characterization of BAV.VII.ST
3.7. Purification and Identification of Cellular/Viral Proteins Interacting with Protein VII
3.8. Interaction between Proteins VII-IVa2 and Proteins VII-VIII
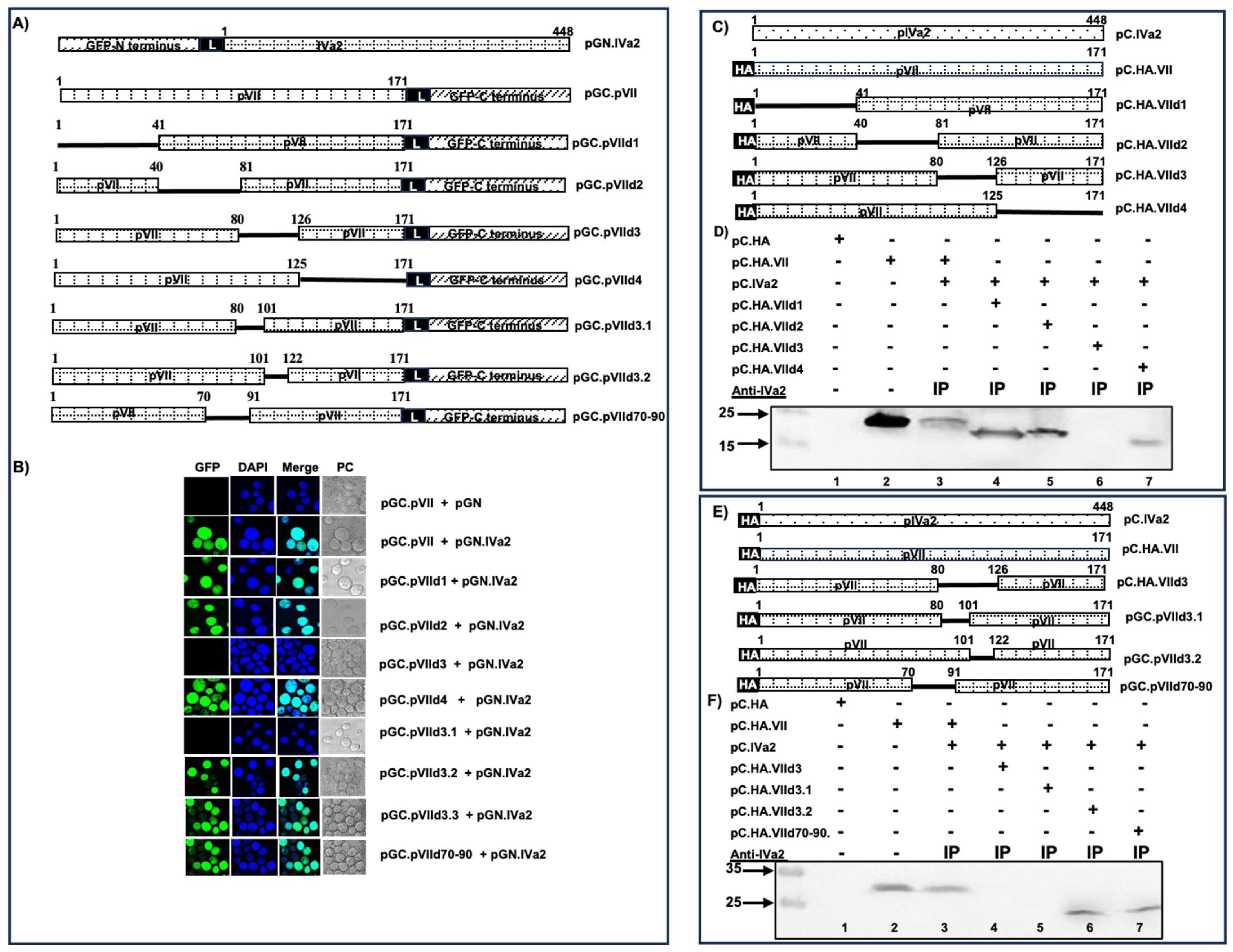
3.9. Identification of Protein VII Domain Interacting with IVa2
3.10. Identification of Protein VII Domain Interacting with Protein VIII
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reddy, P.S.; Idamakanti, N.; Zakhartchouk, A.N.; Baxi, M.K.; Lee, J.B.; Pyne, C.; Babiuk, L.A.; Tikoo, S.K. Nucleotide sequence, genome organization, and transcription map of bovine adenovirus type 3. J. Virol. 1998, 72, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Nemerow, G.R. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11715–11720. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Tikoo, S.K. Nuclear and Nucleolar localization of bovine adenovirus-3 protein V. Front. Microbiol. 2020, 11, 579593. [Google Scholar] [CrossRef] [PubMed]
- Said, A.; Wang, W.; Woldermariam, T.; Tikoo, S.K. Domains of bovine adenovirus-3 protein 22K involved in interacting with viral protein 52K and cellular importins alpha-5/alpha-7. Virology 2018, 522, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Woldemariam, T.; Wang, W.; Said, A.; Tikoo, S.K. Regions of bovine adenovirus-3 IVa2 involved in nuclear/nucleolar localization and interaction with pV. Virology 2020, 546, 25–37. [Google Scholar] [CrossRef]
- Wodrich, H.; Guan, T.; Cingolani, T.; Von Seggern, D.; Nemerow, G.; Gerace, L. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 2003, 22, 6245–6255. [Google Scholar] [CrossRef] [PubMed]
- Wodrich, H.; Cassany, A.; D’Angelo, M.A.; Guan, T.; Nemerow, G.; Gerace, L. Adenovirus core protein pVII is translocated into the nucleus by multiple import receptor pathways. J. Virol. 2006, 80, 9608–9618. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.O.; Dundr, M. The moving parts of the nucleolus. Histochem. Cell Biol. 2005, 123, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.A.; Weber, J. Structure of adenovirus chromatin. Biochim. Biophys. Acta 1982, 696, 76–86. [Google Scholar] [CrossRef]
- Lischwe, M.A.; Sung, M.T. A histone-like protein from adenovirus chromatin. Nature 1977, 267, 552–554. [Google Scholar] [CrossRef]
- Kulanayake, S.; Tikoo, S.K. Adenovirus Core Proteins: Structure and Function. Viruses 2021, 13, 388. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Osheim, Y.N.; Xue, Y.; Emanuel, M.R.; Lewis, P.W.; Bankovich, A.; Beyer, A.L.; Engel, D.A. Adenovirus protein VII condenses DNA, represses transcription, and associates with transcriptional activator E1A. J. Virol. 2004, 78, 6459–6468. [Google Scholar] [CrossRef] [PubMed]
- Puntener, D.; Engelke, M.F.; Ruzsics, Z.; Strunze, S.; Wilhelm, C.; Greber, U.F. Stepwise loss of fluorescent core protein V from human adenovirus during entry into cells. J. Virol. 2011, 85, 481–496. [Google Scholar] [CrossRef] [PubMed]
- Ostapchuk, P.; Suomalainen, M.; Zheng, Y.; Boucke, K.; Greber, U.F.; Hearing, P. The adenovirus major core protein VII is dispensable for virion assembly but is essential for lytic infection. PLoS Pathog. 2017, 13, e1006455. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.J.; Benko, M.; Harrach, B. Genetic content and evolution of adenoviruses. J. Gen. Virol. 2003, 84 Pt 11, 2895–2908. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.H.; Rao, X.M.; McMasters, K.M.; Zhou, H.S. Molecular basis for viral selective replication in cancer cells: Activation of CDK2 by adenovirus induced cyclin E. PLoS ONE 2013, 8, e57340. [Google Scholar] [CrossRef]
- Makadiya, N.; Gaba, A.; Tikoo, S.K. Cleavage of bovine adenovirus type 3 non-structural 100K protein by protease is required for nuclear localization in infected cells but is not essential for virus replication. J. Gen. Virol. 2015, 96, 2749–2763. [Google Scholar] [CrossRef]
- Stracker, T.H.; Lee, D.V.; Carson, C.T.; Araujo, F.D.; Ornelles, D.A.; Weitzman, M.D. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 2005, 79, 6664–6673. [Google Scholar] [CrossRef]
- Papp, Z.; Middleton, D.M.; Mittal, S.K.; Babiuk, L.A.; Baca-Estrada, M.E. Mucosal immunization with recombinant adenoviruses: Induction of immunity and protection of cotton rats against respiratory bovine herpesvirus type 1 infection. J. Gen. Virol. 1997, 78, 2933–2943. [Google Scholar] [CrossRef]
- Du, E.; Tikoo, S.K. Efficient replication and generation of recombinant bovine adenovirus-3 in nonbovine cotton rat lung cells expressing I-SceI endonuclease. J. Gene Med. 2010, 12, 840–847. [Google Scholar] [CrossRef]
- Anand, S.K.; Gaba, A.; Singh, J.; Tikoo, S.K. Bovine adenovirus 3 core protein precursor pVII localizes to mitochondria, and modulates ATP synthesis, mitochondrial Ca2+ and mitochondrial membrane potential. J. Gen. Virol. 2014, 95 Pt 2, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Reddy, P.S.; Babiuk, L.A.; Tikoo, S.K. Bovine adenovirus type 3 E1B(small) protein is essential for growth in bovine fibroblast cells. Virology 2001, 288, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Ayalew, L.E.; Gaba, A.; Kumar, P.; Tikoo, S.K. Conserved regions of bovine adenovirus-3 pVIII contain functional domains involved in nuclear localization and packaging in mature infectious virions. J. Gen. Virol. 2014, 95, 1743–1754. [Google Scholar] [CrossRef] [PubMed]
- Depping, R.; Steinhoff, A.; Schindler, S.G.; Friedrich, B.; Fagerlund, R.; Metzen, E.; Hartmann, E.; Kohler, M. Nuclear translocation of hypoxia-inducible factors (HIFs): Involvement of the classical importib α/β pathway. Biochim. Acta 2008, 1783, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Lin, R.I.; Tarn, W.Y. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Nat. Acad. Sci. USA 2001, 98, 10154–10159. [Google Scholar] [CrossRef] [PubMed]
- Kulshreshtha, V.; Babiuk, L.A.; Tikoo, S.K. Role of bovine adenovirus-3 33K protein in viral replication. Virology 2004, 323, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.; Armon-Omer, A.; Rosenbluh, J.; Melamed-Book, N.; Graessmann, A.; Waigmann, E.; Loyter, A. Inhibition of HIV-1 integrase nuclear import and replication by a peptide bearing integrase putative nuclear localization signal. Retrovirology 2009, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X. The Role of Bovine Adenovirus-3 Protein V (pV) in Virus Replication. Ph.D. Thesis, Department of Veterinary Microbiology, University of Saskatchewan, Saskatoon, SK, Canada, 2016. [Google Scholar]
- Kulshreshtha, V.; Tikoo, S.K. Interaction of bovine adenovirus-3 33K protein with other viral proteins. Virology 2008, 381, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Nat. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef]
- Yachdav, G.; Kloppmann, E.; Kajan, L.; Hecht, M.; Goldberg, T.; Hamp, T.; Honigschmid, P.; Schafferhans, A.; Roos, M.; Bernhofer, M.; et al. PredictProtein—An open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014, 42, 337–343. [Google Scholar] [CrossRef]
- Wu, Q.; Tikoo, S.K. Altered tropism of recombinant bovine adenovirus type-3 expressing chimeric fiber. Virus Res. 2004, 99, 9–15. [Google Scholar] [CrossRef]
- Field, J.; Nikawa, J.; Broek, D.; MacDonald, B.; Rodgers, L.; Wilson, I.A.; Lerner, R.A.; Wigler, M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 1988, 8, 2159–2165. [Google Scholar]
- Gaba, A.; Ayalew, L.; Makadiya, N.; Tikoo, S. Proteolytic Cleavage of Bovine Adenovirus 3-Encoded pVIII. J. Virol. 2017, 91, e00211-17. [Google Scholar] [CrossRef]
- Arnberg, N. Adenovirus receptors: Implications for tropism, treatment and targeting. Rev. Med. Virol. 2009, 19, 165–178. [Google Scholar] [CrossRef]
- Greber, U.F.; Willetts, M.; Webster, P.; Helenius, A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 1993, 75, 477–486. [Google Scholar] [CrossRef]
- Russell, W.C. Adenoviruses: Update on structure and function. J. Gen. Virol. 2009, 90, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Vellinga, J.; Van der Heijdt, S.; Hoeben, R.C. The adenovirus capsid: Major progress in minor proteins. J. Gen. Virol. 2005, 86, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.M. Adenovirus endopeptidase and its role in virus infection. Curr. Top. Microbiol. Immunol. 1995, 199 Pt 1, 227–235. [Google Scholar]
- Dai, X.; Wu, L.; Sun, R.; Zhou, Z.H. Atomic Structures of Minor Proteins VI and VII in Human Adenovirus. J. Virol. 2017, 91, e00850-17. [Google Scholar] [CrossRef]
- Perez-Vargas, J.; Vaughan, R.C.; Houser, C.; Hastie, K.M.; Kao, C.C.; Nemerow, G.R. Isolation and characterization of the DNA and protein binding activities of adenovirus core protein V. J. Virol. 2014, 88, 9287–9296. [Google Scholar] [CrossRef]
- Reddy, V.S.; Barry, M.A. Structural Organization and Protein-Protein Interactions in Human Adenovirus Capsid. Subcell. Biochem. 2021, 96, 503–518. [Google Scholar] [CrossRef] [PubMed]
- Imperiale, M.J.; Akusjnarvi, G.; Leppard, K.N. Post-Transcriptional control of adenovirus gene expression. Curr. Top. Microbiol. Immunol. 1995, 199 Pt 2, 139–171. [Google Scholar] [PubMed]
- Karen, K.A.; Hearing, P. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J. Virol. 2011, 85, 4135–4142. [Google Scholar] [CrossRef] [PubMed]
- Weber, J. Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J. Virol. 1976, 17, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Mangel, W.F.; San Martin, C. Structure, function and dynamics in adenovirus maturation. Viruses 2014, 6, 4536–4570. [Google Scholar] [CrossRef] [PubMed]
- Nemerow, G.R.; Stewart, P.L.; Reddy, V.S. Structure of human adenovirus. Curr. Opin. Virol. 2012, 2, 115–121. [Google Scholar] [CrossRef]
- Boudin, M.L.; D’Halluin, J.C.; Cousin, C.; Boulanger, P. Human adenovirus type 2 protein IIIa. II. Maturation and encapsidation. Virology 1980, 101, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Arcos, R. Interaction of the adenovirus major core protein precursor, pVII, with the viral DNA packaging machinery. Virology 2005, 334, 194–202. [Google Scholar] [CrossRef]
- Cheng, L.; Huang, X.; Li, X.; Xiong, W. Cryo-EM structures of two bovine adenovirus type 3 intermediates. Virology 2014, 450–451, 174–181. [Google Scholar] [CrossRef]
- Gallardo, J.; Pérez-Illana, M.; Martín-González, N.; San Martín, C. Adenovirus structure: What is new? Int. J. Mol. Sci. 2021, 22, 5240. [Google Scholar] [CrossRef]
- Hackenbrack, N.; Rogers, M.B.; Ashley, R.E.; Keel, M.K.; Kubiski, S.V.; Bryan, J.A.; Ghedin, E.; Holmes, E.C.; Hafenstein, S.L.; Allison, A.B. Evolution and cryo-electron microscopy capsid structure of a North-American bat adenovirus and its relationship to other mastadenoviruses. J. Virol. 2017, 91, e01504-16. [Google Scholar] [CrossRef] [PubMed]
- Marabini, R.; Condezo, G.N.; Krupovic, M.; Menendez-Conejero, R.; Gomez-Blanco, J.; San, M.C. Near-atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins. Sci. Adv. 2021, 7, eabe6008. [Google Scholar] [CrossRef] [PubMed]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [PubMed]
- Christophe, D.; Christophe-Hobertus, C.; Pichon, B. Nuclear targeting of proteins: How many different signals? Cell Signal 2000, 12, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Schaley, J.E.; Polonskaia, M.; Hearing, P. The Adenovirus E4-6/7 Protein Directs Nuclear Localization of E2F-4 via an Arginine-Rich Motif. J. Virol. 2005, 79, 2301–2308. [Google Scholar] [CrossRef] [PubMed]
- Al-Wassiti, H.A.; Thomas, D.R.; Wagstaff, K.M.; Fabb, S.A.; Jans, D.A.; Johnston, A.P. Adenovirus Terminal Protein Contains a Bipartite Nuclear Localisation Signal Essential for Its Import into the Nucleus. Int. J. Mol. Sci. 2021, 22, 3310. [Google Scholar] [CrossRef] [PubMed]
- Paterson, C.P.; Ayalew, L.E.; Tikoo, S.K. Mapping of nuclear import signal and importin alpha3 binding regions of 52K protein of bovine adenovirus-3. Virology 2012, 432, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Kulshreshtha, V.; Ayalew, L.E.; Islam, A.; Tikoo, S.K. Conserved arginines of bovine adenovirus-3 33K protein are important for transportin-3 mediated transport and virus replication. PLoS ONE 2014, 9, e101216. [Google Scholar] [CrossRef] [PubMed]
- Reed, M.L.; Dove, B.K.; Jackson, R.M.; Collins, R.; Brooks, G.; Hiscox, J.A. Delineation and modelling of a nucleolar retention signal in the coronavirus nucleocapsid protein. Traffic 2006, 7, 833–848. [Google Scholar] [CrossRef]
- Cheng, G.; Brett, M.E.; He, B. Signals that dictate nuclear, nucleolar, and cytoplasmic shuttling of the gamma (1)34.5 protein of herpes simplex virus type 1. J. Virol. 2002, 76, 9434–9445. [Google Scholar] [CrossRef]
- Cros, J.F.; Garcia-Sastre, A.; Palese, P. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 2005, 6, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Imperiale, M.J. Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol. 2003, 77, 3586–3594. [Google Scholar] [CrossRef] [PubMed]
- Martin-Gonzalez, N.; Hernando-Perez, M.; Condezo, G.N.; Perez-Illana, M.; Siber, A.; Reguera, D.; Ostapchuk, P.; Hearing, P.; San Martin, C.; de Pablo, P.J. Adenovirus major core protein condenses DNA in clusters and bundles, modulating genome release and capsid internal pressure. Nucleic Acids Res. 2019, 47, 9231–9242. [Google Scholar] [CrossRef] [PubMed]
- Perez-Berna, A.J.; Marion, S.; Chichon, F.J.; Fernandez, J.J.; Winkler, D.C.; Carrascosa, J.L.; Steven, A.C.; Siber, A.; San Martin, C. Distribution of DNA-condensing protein complexes in the adenovirus core. Nucleic Acids Res. 2015, 43, 4274–4283. [Google Scholar] [CrossRef] [PubMed]
- Rohn, K.; Prusas, C.; Monreal, G.; Hess, M. Identification and characterization of penton base and pVIII protein of egg drop syndrome virus. Virus Res. 1997, 47, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Perez, M.; Martin-Gonzalez, N.; Perez-Illana, M.; Suomalainen, M.; Condezo, G.N.; Ostapchuk, P.; Gallardo, J.; Menendez, M.; Greber, U.F.; Hearing, P.; et al. Dynamic competition for hexon binding between core protein VII and lytic protein VI promotes adenovirus maturation and entry. Proc. Natl. Acad. Sci. USA 2020, 117, 13699–13707. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Q.; Babiss, L.E.; Volkert, F.C.; Young, C.S.; Ginsberg, H.S. A thermolabile mutant of adenovirus 5 resulting from a substitution mutation in the protein VIII gene. J. Virol. 1985, 53, 920–925. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.A.; Rose, R.E. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res 1988, 16, 358. [Google Scholar] [CrossRef]
- Zakhartchouk, A.; Connors, W.; Van Kessel, A.; Tikoo, S.K. Bovine adenovirus type 3 containing heterologous protein in the C-terminus of minor capsid protein IX. Virology 2004, 320, 291–300. [Google Scholar] [CrossRef]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulanayake, S.; Dar, F.; Tikoo, S.K. Regions of Bovine Adenovirus-3 Protein VII Involved in Interactions with Viral and Cellular Proteins. Viruses 2024, 16, 732. https://doi.org/10.3390/v16050732
Kulanayake S, Dar F, Tikoo SK. Regions of Bovine Adenovirus-3 Protein VII Involved in Interactions with Viral and Cellular Proteins. Viruses. 2024; 16(5):732. https://doi.org/10.3390/v16050732
Chicago/Turabian StyleKulanayake, Shermila, Faryal Dar, and Suresh K. Tikoo. 2024. "Regions of Bovine Adenovirus-3 Protein VII Involved in Interactions with Viral and Cellular Proteins" Viruses 16, no. 5: 732. https://doi.org/10.3390/v16050732




