Integrated LSPR Biosensing Signal Processing Strategy and Visualization Implementation
Abstract
:1. Introduction
2. LSPR Sensing Principles on the Integrated Visual Software
2.1. Comparison of Different LSPR Sensing Parameters
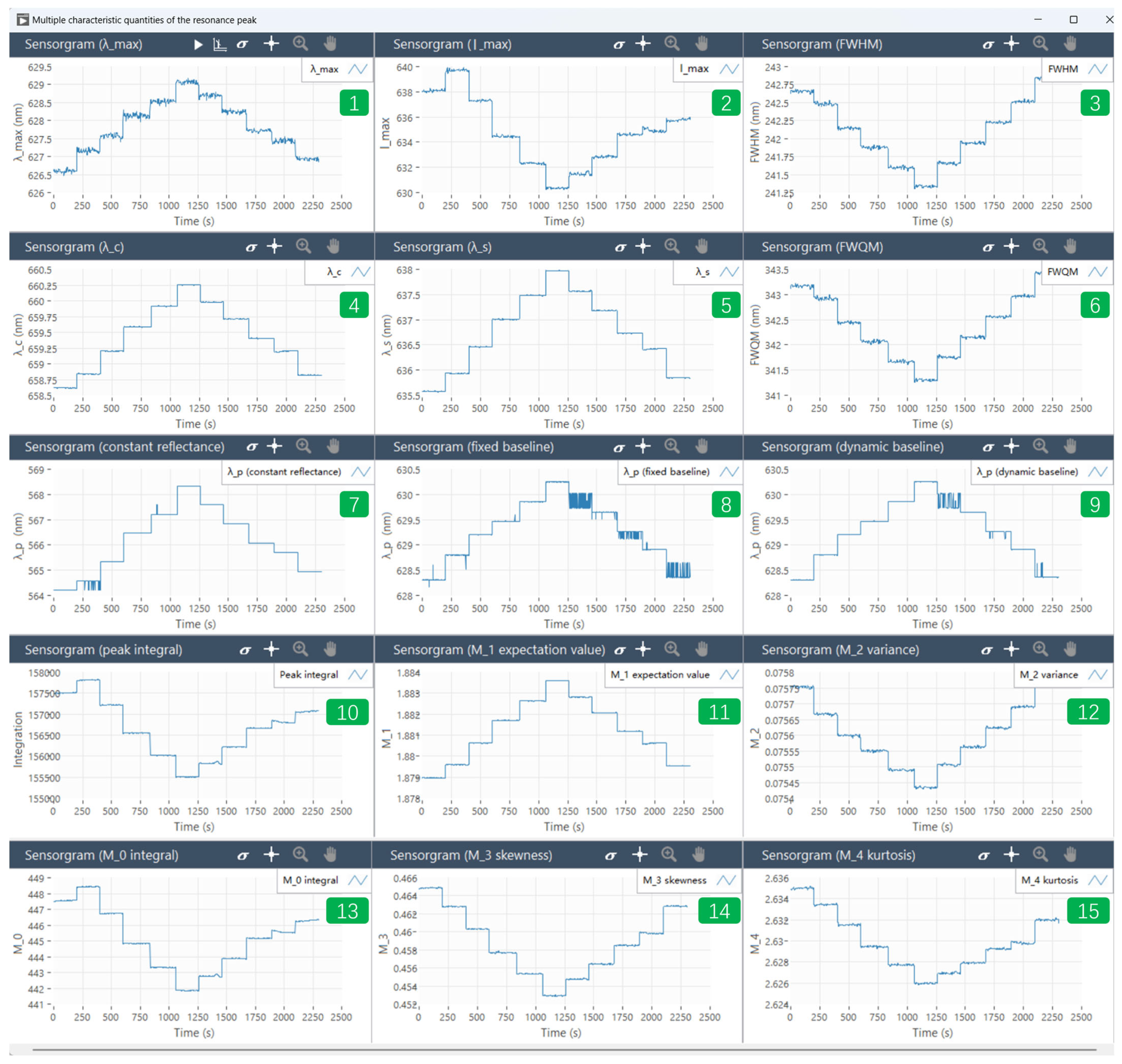
2.2. Multiple Resonance Peak Characterization Algorithms Integrated in the Visual Software
2.3. LSPR Biosensor Signal Acquisition through Visual Software
- (1)
- System control. The software controls multiple devices and implements various algorithms for data processing. Therefore, in cases where multiple devices are involved, the system needs to manage the connected devices and choose from a variety of processing modes. In addition, the individual modules need to be managed and controlled.
- (2)
- Spectra acquisition. According to the communication protocol, the software uses the serial port to transmit the control command to the sensor and realizes the communication with the sensor to collect and transmit spectral data.
- (3)
- Spectra control. The spectra collected from the sensor are processed. According to the different needs of users, there will be different processing methods, mainly including the calculation of spectral characteristic parameters, background removal, spectral superposition, etc.
- (4)
- Data manipulation. During system operation, data undergo meticulous management, encompassing tasks.
- (5)
- Data processing. The system’s core functions revolve around data processing and analysis, encompassing advanced processing of acquired spectra, extraction of relevant features, and subsequent utilization of these features for thorough analysis. It reads data from the sensor, conducting calculations based on the user-selected mode to facilitate functions like noise reduction, spectral baseline subtraction, identification of feature peaks, and comprehensive spectral analysis.
- (6)
- Interaction. The interactive interface acts as the channel for human–computer interaction and stands as the exclusive component of the system accessible to users. Consisting of a control interface and a spectra interface, this user interaction interface predominantly functions in observer mode, overseeing distinct controls and triggering diverse functions through an integration with the underlying model.
3. Visualization Implements of Integrated LSPR Signal Processing Software
4. Results and Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- He, J.; Wu, S.; Chen, W.; Kim, A.; Yang, W.; Wang, C.; Gu, Z.; Shen, J.; Dai, S.; Chen, W.; et al. Calligraphy of Nanoplasmonic Bioink-Based Multiplex Immunosensor for Precision Immune Monitoring and Modulation. ACS Appl. Mater. Interfaces 2023, 15, 50047–50057. [Google Scholar] [CrossRef]
- Xu, T.; Geng, Z. Strategies to Improve Performances of LSPR Biosensing: Structure, Materials, and Interface Modification. Biosens. Bioelectron. 2021, 174, 112850. [Google Scholar] [CrossRef]
- Proença, M.; Rodrigues, M.S.; Borges, J.; Vaz, F. Optimization of Au:CuO Nanocomposite Thin Films for Gas Sensing with High-Resolution Localized Surface Plasmon Resonance Spectroscopy. Anal. Chem. 2020, 92, 4349–4356. [Google Scholar] [CrossRef]
- Palani, S.; Kenison, J.P.; Sabuncu, S.; Huang, T.; Civitci, F.; Esener, S.; Nan, X. Multispectral Localized Surface Plasmon Resonance (msLSPR) Reveals and Overcomes Spectral and Sensing Heterogeneities of Single Gold Nanoparticles. ACS Nano 2023, 17, 2266–2278. [Google Scholar] [CrossRef]
- Huang, T.; Wang, J.; Duan, F.; Jiang, J.; Fu, X.; Xu, X. A Simple Algorithm for the Implementation of Second-Order-Polynomial Based Peak-Tracking Methods. Opt. Fiber Technol. 2019, 47, 192–196. [Google Scholar] [CrossRef]
- Yang, G.; Dai, J.; Liu, X.; Chen, M.; Wu, X. Spectral Feature Extraction Based on Continuous Wavelet Transform and Image Segmentation for Peak Detection. Anal. Methods 2020, 12, 169–178. [Google Scholar] [CrossRef]
- Gul, M.U.; Kadir, K.; Azman, H.K.; Iqbal, S. Detection of R-Peaks Using Single-Scale Wavelet Transform. In Proceedings of the 2019 13th International Conference on Mathematics, Actuarial Science, Computer Science and Statistics (MACS), Karachi, Pakistan, 14–15 December 2019; pp. 1–5. [Google Scholar]
- Ma, K.; Liu, L.; Zhang, P.; He, Y.; Peng, Q. Optimization of Angle-Pixel Resolution for Angular Plasmonic Biosensors. Sens. Actuators B Chem. 2019, 283, 188–197. [Google Scholar] [CrossRef]
- Wang, G.; Shi, J.; Zhang, Q.; Wang, R.; Huang, L. Resolution Enhancement of Angular Plasmonic Biochemical Sensors via Optimizing Centroid Algorithm. Chemom. Intell. Lab. Syst. 2022, 223, 104531. [Google Scholar] [CrossRef]
- Jeon, J.; Uthaman, S.; Lee, J.; Hwang, H.; Kim, G.; Yoo, P.J.; Hammock, B.D.; Kim, C.S.; Park, Y.-S.; Park, I.-K. In-Direct Localized Surface Plasmon Resonance (LSPR)-Based Nanosensors for Highly Sensitive and Rapid Detection of Cortisol. Sens. Actuators B Chem. 2018, 266, 710–716. [Google Scholar] [CrossRef]
- Baihaqi, M.Y.; Lumoindong, C.W.D.; Vincent, V. Simulasi Perbandingan Filter Savitzky Golay Dan Filter Low Pass Butterworth Pada Orde Ketiga Sebagai Pembatal Kebisingan. KONSTELASI Konvergensi Teknol. Sist. Inf. 2021, 1, 226–232. [Google Scholar] [CrossRef]
- Zhang, G.; Hao, H.; Wang, Y.; Jiang, Y.; Shi, J.; Yu, J.; Cui, X.; Li, J.; Zhou, S.; Yu, B. Optimized Adaptive Savitzky-Golay Filtering Algorithm Based on Deep Learning Network for Absorption Spectroscopy. Spectrochim. Acta Part A 2021, 263, 120187. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Z.; Wang, J.; Wang, J. Time–Frequency Analysis for Bearing Fault Diagnosis Using Multiple Q-Factor Gabor Wavelets. ISA Trans. 2019, 87, 225–234. [Google Scholar] [CrossRef]
- Altenhof, A.R.; Mason, H.; Schurko, R.W. DESPERATE: A Python Library for Processing and Denoising NMR Spectra. J. Magn. Reson. 2023, 346, 107320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Qi, W.; Cen, Y.; Lin, H.; Wang, N. Denoising Vegetation Spectra by Combining Mathematical-Morphology and Wavelet-Transform-Based Filters. J. Appl. Remote Sens. 2019, 13, 1. [Google Scholar] [CrossRef]
- Krämer, S.D.; Wöhrle, J.; Rath, C.; Roth, G. Anabel: An Online Tool for the Real-Time Kinetic Analysis of Binding Events. Bioinf. Biol. Insights 2019, 13, 117793221882138. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.B.; Rodrigues, E.P.; Pereira, H.A. Sim-SPR: An Open-Source Surface Plasmon Resonance Simulator for Academic and Industrial Purposes. Plasmonics 2019, 14, 1699–1709. [Google Scholar] [CrossRef]
- Dahl, G.; Steigele, S.; Hillertz, P.; Tigerström, A.; Egnéus, A.; Mehrle, A.; Ginkel, M.; Edfeldt, F.; Holdgate, G.; O’Connell, N.; et al. Unified Software Solution for Efficient SPR Data Analysis in Drug Research. SLAS Discov. 2017, 22, 203–211. [Google Scholar] [CrossRef]
- Rodrigues, M.S.; Pereira, R.M.S.; Vasilevskiy, M.I.; Borges, J.; Vaz, F. NANOPTICS: In-Depth Analysis of NANomaterials for OPTICal Localized Surface Plasmon Resonance Sensing. SoftwareX 2020, 12, 100522. [Google Scholar] [CrossRef]
- Muri, H.I.; Bano, A.; Hjelme, D.R. A Single-Point, Multiparameter, Fiber Optic Sensor Based on a Combination of Interferometry and LSPR. J. Light. Technol. 2018, 36, 1159–1167. [Google Scholar] [CrossRef]
- Paul, D.; Biswas, R. Highly Sensitive LSPR Based Photonic Crystal Fiber Sensor with Embodiment of Nanospheres in Different Material Domain. Opt. Laser Technol. 2018, 101, 379–387. [Google Scholar] [CrossRef]
- Santos, C.; Dias, C. Note on the Coefficient of Variation Properties. Braz. Electron. J. Math. 2021, 2, 101–111. [Google Scholar] [CrossRef]
- Çataltaş, Ö.; Tutuncu, K. A Review of Data Analysis Techniques Used in Near-Infrared Spectroscopy. Eur. J. Sci. Technol. 2021, 25, 475–484. [Google Scholar] [CrossRef]
- Chinowsky, T.M.; Jung, L.S.; Yee, S.S. Optimal Linear Data Analysis for Surface Plasmon Resonance Biosensors. Sens. Actuators B Chem. 1999, 54, 89–97. [Google Scholar] [CrossRef]
- Thirstrup, C.; Zong, W. Data Analysis for Surface Plasmon Resonance Sensors Using Dynamic Baseline Algorithm. Sens. Actuators B Chem. 2005, 106, 796–802. [Google Scholar] [CrossRef]
- Dos Santos, P.S.S.; Mendes, J.P.; Dias, B.; Pérez-Juste, J.; De Almeida, J.M.M.M.; Pastoriza-Santos, I.; Coelho, L.C.C. Spectral Analysis Methods for Improved Resolution and Sensitivity: Enhancing SPR and LSPR Optical Fiber Sensing. Sensors 2023, 23, 1666. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Lin, X.; Shao, X. A Wavelet Transform and Its Application to Spectroscopic Analysis. Appl. Spectrosc. Rev. 2002, 37, 429–450. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Milojković-Opsenica, D.; Ristivojević, P.; Andrić, F.; Trifković, J. Planar Chromatographic Systems in Pattern Recognition and Fingerprint Analysis. Chromatographia 2013, 76, 1239–1247. [Google Scholar] [CrossRef]
- Leung, A.K.; Chau, F.; Gao, J. A Review on Applications of Wavelet Transform Techniques in Chemical Analysis: 1989–1997. Chemom. Intell. Lab. Syst. 1998, 43, 165–184. [Google Scholar] [CrossRef]
- Kwiatkowski, A.; Gnyba, M.; Smulko, J.; Wierzba, P. Algorithms of Chemicals Detection Using Raman Spectra. Metrol. Meas. Syst. 2010, 17, 549–559. [Google Scholar] [CrossRef]
- Zhang, F.; Tang, X.; Tong, A.; Wang, B.; Wang, J. An Automatic Baseline Correction Method Based on the Penalized Least Squares Method. Sensors 2020, 20, 2015. [Google Scholar] [CrossRef] [PubMed]
- Ruckstuhl, A.F.; Jacobson, M.P.; Field, R.W.; Dodd, J.A. Baseline Subtraction Using Robust Local Regression Estimation. J. Quant. Spectrosc. Radiat. Transfer 2001, 68, 179–193. [Google Scholar] [CrossRef]
- Jiang, X.Q.; Blunt, L.; Stout, K.J. Development of a Lifting Wavelet Representation for Surface Characterization. Proc. R. Soc. Lond. A Math. Phys. Eng. Sci. 2000, 456, 2283–2313. [Google Scholar] [CrossRef]
- Hoang, V.D. Wavelet-Based Spectral Analysis. TrAC Trends Anal. Chem. 2014, 62, 144–153. [Google Scholar] [CrossRef]
- Perez-Pueyo, R.; Soneira, M.J.; Ruiz-Moreno, S. Morphology-Based Automated Baseline Removal for Raman Spectra of Artistic Pigments. Appl. Spectrosc. 2010, 64, 595–600. [Google Scholar] [CrossRef]
- Zhang, Z.-M.; Chen, S.; Liang, Y.-Z.; Liu, Z.-X.; Zhang, Q.-M.; Ding, L.-X.; Ye, F.; Zhou, H. An Intelligent Background-Correction Algorithm for Highly Fluorescent Samples in Raman Spectroscopy: Background-Correction Algorithm for Highly Fluorescent Samples in Raman Spectroscopy. J. Raman Spectrosc. 2010, 41, 659–669. [Google Scholar] [CrossRef]








| Characteristic Parameters | Standard Derivation | Variance | Coefficient of Variation |
|---|---|---|---|
| 4.55770 × 10−2 | 2.07727 × 10−3 | 7.27371 × 10−3 | |
| 6.28609 × 10−2 | 3.95149 × 10−3 | 9.85157 × 10−3 | |
| Peak area integral | 4.85507 | 23.5717 | 3.08245 × 10−3 |
| 3.77299 × 10−3 | 1.42355 × 10−5 | 5.72865 × 10−4 | |
| 1.52819 × 10−2 | 2.33537 × 10−4 | 6.29772 × 10−3 | |
| 2.10774 × 10−2 | 4.44257 × 10−4 | 6.14196 × 10−3 | |
| 5.04019 × 10−3 | 2.54035 × 10−5 | 7.93009 × 10−4 | |
| Constant reflectance | 1.14081 × 10−13 | 1.30145 × 10−26 | 2.02198 × 10−14 |
| Fixed baseline | 2.55323 × 10−3 | 6.51897 × 10−6 | 4.06364 × 10−4 |
| Dynamic baseline | 1.50999 × 10−3 | 2.28008 × 10−6 | 2.40326 × 10−4 |
| Integral | 1.74456 × 10−2 | 3.04349 × 10−4 | 3.89805 × 10−3 |
| Expectation value | 8.78245 × 10−6 | 7.71315 × 10−11 | 4.67409 × 10−4 |
| Variance | 2.91788 × 10−6 | 8.51404 × 10−12 | 3.85185 × 10−3 |
| Skewness | 3.50356 × 10−5 | 1.22749 × 10−9 | 7.53655 × 10−3 |
| Kurtosis | 3.49702 × 10−5 | 1.22291 × 10−9 | 1.32713 × 10−3 |
| Characteristic Parameters | S (nm/RIU) | FOM (RIU−1) | (Linear Fit, nm) | R (RIU) | LOD (RIU2/nm) |
|---|---|---|---|---|---|
| 54.5156 | 0.224660 | 1.58958 × 10−3 | 2.91583 × 10−5 | 5.34861 × 10−7 | |
| 37.8449 | 0.155959 | 1.27462 × 10−3 | 3.36801 × 10−5 | 8.89952 × 10−7 | |
| 30.5045 | 0.125709 | 7.68332 × 10−4 | 2.51875 × 10−5 | 8.25696 × 10−7 | |
| 43.1601 | 0.177863 | 1.56704 × 10−3 | 3.63075 × 10−5 | 8.41229 × 10−7 | |
| 55.0763 | 0.226970 | 1.68602 × 10−3 | 3.06124 × 10−5 | 5.55818 × 10−7 | |
| Fixed baseline | 42.1106 | 0.173538 | 2.63576 × 10−3 | 6.25914 × 10−5 | 1.48636 × 10−6 |
| Dynamic baseline | 42.1075 | 0.173526 | 2.64558 × 10−3 | 6.28291 × 10−5 | 1.49211 × 10−6 |
| Constant reflection | 95.2644 | 0.392587 | 3.55440 × 10−2 | 3.73109 × 10−4 | 3.91657 × 10−6 |
| Characteristic Parameters | S (A/RIU) | FOM (RIU−1) | (Linear Fit, A) | R (RIU) | LOD (RIU2/A) |
| 21.3569 | 8.80116 × 10−2 | 1.33507 × 10−2 | 6.24588 × 10−5 | 2.92203 × 10−5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, M.; Geng, Z. Integrated LSPR Biosensing Signal Processing Strategy and Visualization Implementation. Micromachines 2024, 15, 631. https://doi.org/10.3390/mi15050631
Zhou M, Geng Z. Integrated LSPR Biosensing Signal Processing Strategy and Visualization Implementation. Micromachines. 2024; 15(5):631. https://doi.org/10.3390/mi15050631
Chicago/Turabian StyleZhou, Mixing, and Zhaoxin Geng. 2024. "Integrated LSPR Biosensing Signal Processing Strategy and Visualization Implementation" Micromachines 15, no. 5: 631. https://doi.org/10.3390/mi15050631





