Effects of Transplantation and Microhabitat on Rhizosphere Microbial Communities during the Growth of American Ginseng
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup and Soil Collection
2.2. DNA Extraction, Amplification and Sequence Processing
2.3. Soil Physico-Chemical Properties and Morpho-Physiological Parameters
2.4. Statistical Analysis
3. Results
3.1. Soil and Ginseng Rhizosphere Microbiota Harbor Distinct Communities after Transplanting
3.2. Different Structures and Variation of Soil and Rhizosphere Microbiota in Transplanting Process
3.3. Composition and Co-Occurrence Network of Microbial Communities in Different Microhabitats before and after Transplanting
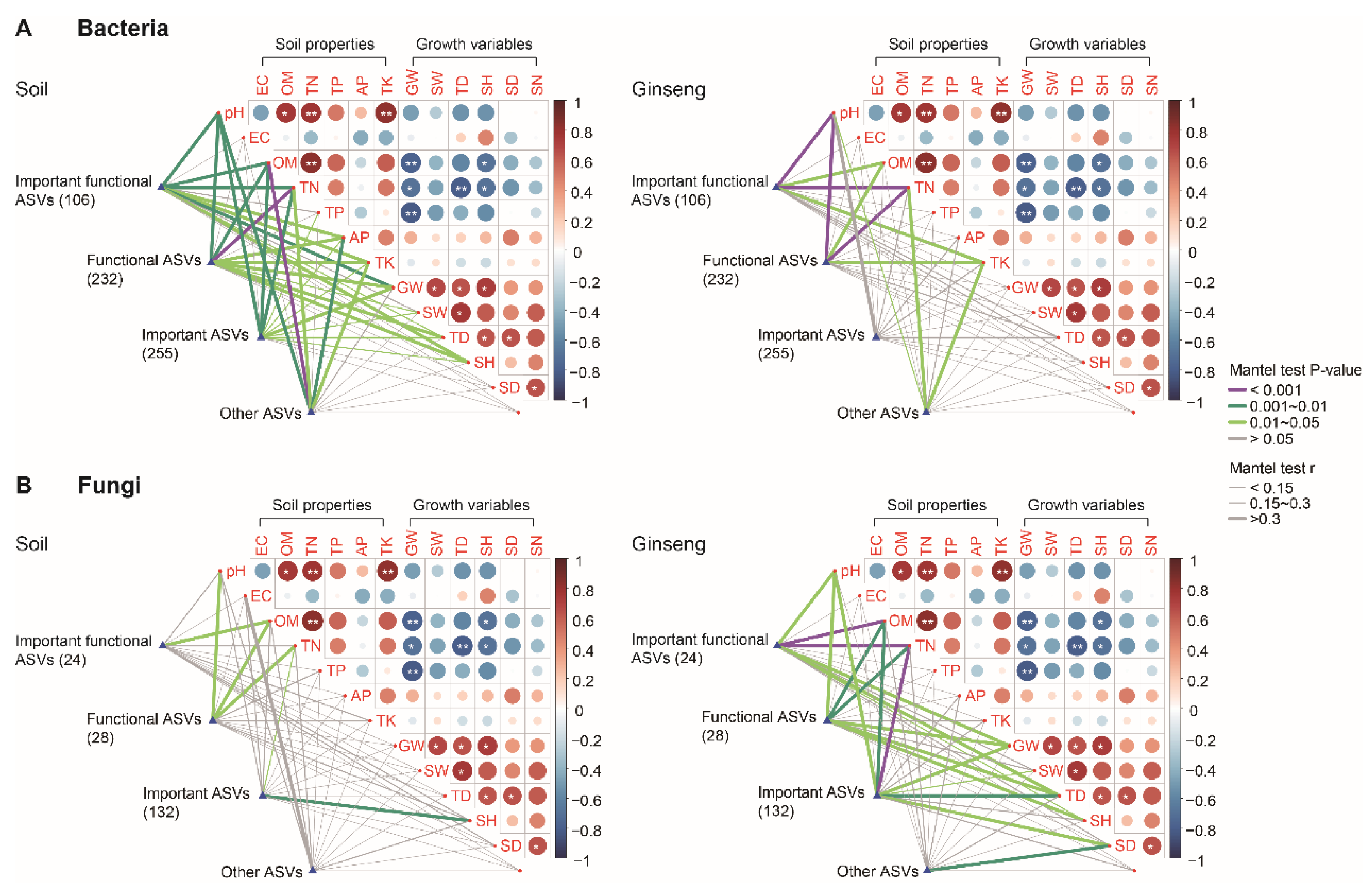
3.4. Association of Microbial Biomarkers and Functional Microorganisms with Soil Properties and Growth of American Ginseng
3.5. Driving Forces for American Ginseng American Growth
4. Discussion
4.1. Response of Soil and Rhizosphere Microhabitat Microbial Communities to Transplanting of American Ginseng
4.2. Microbial Composition Exhibited Distinct Correlation Network Patterns in Different Microhabitats before and after Transplanting
4.3. Effects of Microbial Communities and Soil Properties on Growth Variables in Soil and Ginseng Root Microhabitats
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xue, P.; Yao, Y.; Yang, X.-S.; Feng, J.; Ren, G.-X. Improved antimicrobial effect of ginseng extract by heat transformation. J. Ginseng Res. 2017, 41, 180–187. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.H.; Lee, D.; Lee, H.L.; Kim, C.-E.; Jung, K.; Kang, K.S. Beneficial effects of Panax ginseng for the treatment and prevention of neurodegenerative diseases: Past findings and future directions. J. Ginseng Res. 2018, 42, 239–247. [Google Scholar] [CrossRef]
- Huang, X.; Liu, Y.; Zhang, Y.; Li, S.-P.; Yue, H.; Chen, C.-B.; Liu, S.-Y. Multicomponent assessment and ginsenoside conversions of Panax quinquefolium L. roots before and after steaming by HPLC-MSn. J. Ginseng Res. 2019, 43, 27–37. [Google Scholar] [CrossRef]
- Jiao, X.-L.; Zhang, X.-S.; Lu, X.-H.; Qin, R.; Bi, Y.-M.; Gao, W.-W. Effects of maize rotation on the physicochemical properties and microbial communities of American ginseng cultivated soil. Sci. Rep. 2019, 9, 8615. [Google Scholar] [CrossRef] [Green Version]
- Chang, F.; Jia, F.; Lv, R.; Guan, M.; Jia, Q.; Sun, Y.; Li, Z. Effects of American Ginseng Cultivation on Bacterial Community Structure and Responses of Soil Nutrients in Different Ecological Niches. J. Microbiol. Biotechnol. 2022, 32, 419–429. [Google Scholar] [CrossRef]
- Xiao, C.; Yang, L.; Zhang, L.; Liu, C.; Han, M. Effects of cultivation ages and modes on microbial diversity in the rhizosphere soil of Panax ginseng. J. Ginseng Res. 2016, 40, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Chung, I.-M.; Lee, T.-J.; Oh, Y.-T.; Ghimire, B.K.; Jang, I.-B.; Kim, S.-H. Ginseng authenticity testing by measuring carbon, nitrogen, and sulfur stable isotope compositions that differ based on cultivation land and organic fertilizer type. J. Ginseng Res. 2017, 41, 195–200. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Suo, Y.; Wei, H.; Li, M.; Xie, C.; Wang, L.; Chen, X.; Zhang, Z. Identification and Characterization of 40 Isolated Rehmannia glutinosa MYB Family Genes and Their Expression Profiles in Response to Shading and Continuous Cropping. IJMS 2015, 16, 15009–15030. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Yang, M.; Liu, Y.; Huang, H.; Ye, C.; Zheng, J.; Guo, C.; Hao, M.; He, X.; Zhu, S. Fertilizer N application rate impacts plant-soil feedback in a sanqi production system. Sci. Total Environ. 2018, 633, 796–807. [Google Scholar] [CrossRef]
- Zhang, J.; Fan, S.; Qin, J.; Dai, J.; Zhao, F.; Gao, L.; Lian, X.; Shang, W.; Xu, X.; Hu, X. Changes in the Microbiome in the Soil of an American Ginseng Continuous Plantation. Front. Plant Sci. 2020, 11, 572199. [Google Scholar] [CrossRef]
- Dong, L.; Xu, J.; Zhang, L.; Yang, J.; Liao, B.; Li, X.; Chen, S. High-throughput sequencing technology reveals that continuous cropping of American ginseng results in changes in the microbial community in arable soil. Chin. Med. 2017, 12, 18. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.; Wu, J.; Ji, Q.; Wu, W.; Dong, S.; Yu, J.; Zhang, Q.; Qin, L. Diversity of rhizosphere and endophytic fungi in Atractylodes macrocephala during continuous cropping. PeerJ 2020, 8, e8905. [Google Scholar] [CrossRef] [Green Version]
- DesRochers, N.; Walsh, J.P.; Renaud, J.B.; Seifert, K.A.; Yeung, K.K.-C.; Sumarah, M.W. Metabolomic Profiling of Fungal Pathogens Responsible for Root Rot in American Ginseng. Metabolites 2020, 10, 35. [Google Scholar] [CrossRef] [Green Version]
- Tao, C.; Li, R.; Xiong, W.; Shen, Z.; Liu, S.; Wang, B.; Ruan, Y.; Geisen, S.; Shen, Q.; Kowalchuk, G.A. Bio-organic fertilizers stimulate indigenous soil Pseudomonas populations to enhance plant disease suppression. Microbiome 2020, 8, 137. [Google Scholar] [CrossRef]
- Bennett, J.A.; Klironomos, J. Mechanisms of plant–soil feedback: Interactions among biotic and abiotic drivers. New Phytol. 2019, 222, 91–96. [Google Scholar] [CrossRef] [Green Version]
- Toal, M.E.; Yeomans, C.; Killham, K.; Meharg, A. A review of rhizosphere carbon flow modelling. Plant Soil 2000, 222, 263–281. [Google Scholar] [CrossRef]
- Tong, A.-Z.; Liu, W.; Liu, Q.; Xia, G.-Q.; Zhu, J.-Y. Diversity and composition of the Panax ginseng rhizosphere microbiome in various cultivation modesand ages. BMC Microbiol. 2021, 21, 18. [Google Scholar] [CrossRef]
- Wu, J.; Shi, Z.; Zhu, J.; Cao, A.; Fang, W.; Yan, D.; Wang, Q.; Li, Y. Taxonomic response of bacterial and fungal populations to biofertilizers applied to soil or substrate in greenhouse-grown cucumber. Sci. Rep. 2022, 12, 18522. [Google Scholar] [CrossRef]
- Cao, X.; Liu, S.; Chen, L.; Wang, J.; Xiang, D.; Zhang, J.; Qiao, Z. Effects of different crops on rhizosphere bacterial diversity under immature soil conditions. Arch. Agron. Soil Sci. 2020, 68, 18–30. [Google Scholar] [CrossRef]
- Lazcano, C.; Boyd, E.; Holmes, G.; Hewavitharana, S.; Pasulka, A.; Ivors, K. The rhizosphere microbiome plays a role in the resistance to soil-borne pathogens and nutrient uptake of strawberry cultivars under field conditions. Sci. Rep. 2021, 11, 3188. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, T.; Li, D.; Fu, J.; Liu, S. iMeta: Integrated Meta-Omics for Biology and Environments. iMeta [Internet] 2022; 1. Available online: https://onlinelibrary.wiley.com/doi/10.1002/imt2.15 (accessed on 5 July 2023).
- Zhang, B.; Peng, Y.; Zhang, Z.; Liu, H.; Qi, Y.; Liu, S.; Xiao, P. GAP Production of TCM Herbs in China. Planta Med. 2010, 76, 1948–1955. [Google Scholar] [CrossRef]
- Logue, J.B.; Stedmon, C.A.; Kellerman, A.M.; Nielsen, N.J.; Andersson, A.F.; Laudon, H.; Lindström, E.S.; Kritzberg, E.S. Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. ISME J. 2016, 10, 533–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlsson, I.; Friberg, H.; Steinberg, C.; Persson, P. Fungicide Effects on Fungal Community Composition in the Wheat Phyllosphere. PLoS ONE 2014, 9, e111786. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Valencia, A., editor. Bioinformatics 2018, 34, 2371–2375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacci, G.; Bani, A.; Bazzicalupo, M.; Ceccherini, M.T.; Galardini, M.; Nannipieri, P.; Pietramellara, G.; Mengoni, A. Evaluation of the Performances of Ribosomal Database Project (RDP) Classifier for Taxonomic Assignment of 16S rRNA Metabarcoding Sequences Generated from Illumina-Solexa NGS. J. Genom. 2015, 3, 36–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, B.; Liu, Y.; Sun, X.; Li, S.; Wang, X.; Xiong, K.; Yun, B.; Zhang, H. Effect of various mulches on soil physico—Chemical properties and tree growth (Sophora japonica) in urban tree pits. PLoS ONE 2019, 14, e0210777. [Google Scholar] [CrossRef] [Green Version]
- Fan, K.; Weisenhorn, P.; Gilbert, J.A.; Shi, Y.; Bai, Y.; Chu, H. Soil pH correlates with the co-occurrence and assemblage process of diazotrophic communities in rhizosphere and bulk soils of wheat fields. Soil Biol. Biochem. 2018, 121, 185–192. [Google Scholar] [CrossRef]
- Bradstreet, R.B. Kjeldahl Method for Organic Nitrogen. Anal. Chem. 1954, 26, 185–187. [Google Scholar] [CrossRef]
- Khan, S.A.; Mulvaney, R.L.; Mulvaney, C.S. Accelerated Diffusion Methods for Inorganic-Nitrogen Analysis of Soil Extracts and Water. Soil Sci. Soc. Am. J. 1997, 61, 936–942. [Google Scholar] [CrossRef]
- Murphy, J.A.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Wolf, A.M.; Baker, D.E. Comparisons of soil test phosphorus by Olsen, Bray P1, Mehlich I and Mehlich III methods. Commun. Soil Sci. Plant Anal. 1985, 16, 467–484. [Google Scholar] [CrossRef]
- Attoe, O.J.; Truog, E. Rapid Photometric Determination of Exchangeable Potassium and Sodium. Soil Sci. Soc. Am. J. 1947, 11, 221–226. [Google Scholar] [CrossRef]
- Chen, T.; Liu, Y.; Huang, L. ImageGP: An Easy-to-Use Data Visualization Web Server for Scientific Researchers. iMeta [Internet] 2022; 1. Available online: https://onlinelibrary.wiley.com/doi/10.1002/imt2.5 (accessed on 5 July 2023).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package [Internet] 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 5 July 2023).
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [Green Version]
- Webb, C.O.; Ackerly, D.D.; McPeek, M.A.; Donoghue, M.J. Phylogenies and Community Ecology. Annu. Rev. Ecol. Syst. 2002, 33, 475–505. [Google Scholar] [CrossRef] [Green Version]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef] [Green Version]
- Levins, R. Evolution in changing environments: Some theoretical explorations; Monographs in Population BiologyPrinceton University Press: Princeton, NJ, USA; p. 1968.
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. ICWSM 2009, 3, 361–362. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Diniz-Filho, J.A.; Soares, T.N.; Lima, J.S.; Dobrovolski, R.; Lemes Landeiro, V.; Pires de Campos Telles, M.; Rangel, T.F.; Bini, L.M. Mantel test in population genetics. Genet. Mol. Biol. 2013, 36, 475–485. [Google Scholar] [CrossRef] [Green Version]
- Campbell, B.J.; Yu, L.; Heidelberg, J.F.; Kirchman, D.L. Activity of abundant and rare bacteria in a coastal ocean. Proc. Natl. Acad. Sci. USA 2011, 108, 12776–12781. [Google Scholar] [CrossRef]
- Jiao, S.; Lu, Y. Soil pH and temperature regulate assembly processes of abundant and rare bacterial communities in agricultural ecosystems. Environ. Microbiol. 2020, 22, 1052–1065. [Google Scholar] [CrossRef]
- Tenenhaus, M.; Vinzi, V.E.; Chatelin, Y.-M.; Lauro, C. PLS path modeling. Comput. Stat. Data Anal. 2005, 48, 159–205. [Google Scholar] [CrossRef]
- Li, G.-L.; Wu, M.; Li, P.-F.; Wei, S.-P.; Liu, J.; Jiang, C.-Y.; Liu, M.; Li, Z.-P. Assembly and co-occurrence patterns of rare and abundant bacterial sub-communities in rice rhizosphere soil under short-term nitrogen deep placement. J. Integr. Agric. 2021, 20, 3299–3311. [Google Scholar] [CrossRef]
- Wei, X.; Wang, X.; Cao, P.; Gao, Z.; Chen, A.J.; Han, J. Microbial Community Changes in the Rhizosphere Soil of Healthy and Rusty Panax ginseng and Discovery of Pivotal Fungal Genera Associated with Rusty Roots. BioMed Res. Int. 2020, 2020, 8018525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Wang, Z.; Niu, J.; Dang, K.; Zhang, S.; Wang, S.; Wang, Z. Changes in physicochemical properties, enzymatic activities, and the microbial community of soil significantly influence the continuous cropping of Panax quinquefolius L. (American ginseng). Plant Soil 2021, 463, 427–446. [Google Scholar] [CrossRef]
- Ji, L.; Tian, L.; Nasir, F.; Chang, J.; Chang, C.; Zhang, J.; Li, X.; Tian, C. Impacts of replanting American ginseng on fungal assembly and abundance in response to disease outbreaks. Arch. Microbiol. 2021, 203, 2157–2170. [Google Scholar] [CrossRef]
- Ji, L.; Nasir, F.; Tian, L.; Chang, J.; Sun, Y.; Zhang, J.; Li, X.; Tian, C. Outbreaks of Root Rot Disease in Different Aged American Ginseng Plants Are Associated With Field Microbial Dynamics. Front. Microbiol. 2021, 12, 676880. [Google Scholar] [CrossRef]
- Favela, A.; Bohn, M.O.; Kent, A.D. Maize germplasm chronosequence shows crop breeding history impacts recruitment of the rhizosphere microbiome. ISME J. 2021, 15, 2454–2464. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.D.J.; Neufeld, J.D. Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 2015, 13, 217. [Google Scholar] [CrossRef]
- Dahlstrom, K.M.; McRose, D.L.; Newman, D.K. Keystone metabolites of crop rhizosphere microbiomes. Curr. Biol. 2020, 30, R1131–R1137. [Google Scholar] [CrossRef]
- Yuan, M.M.; Guo, X.; Wu, L.; Zhang, Y.; Xiao, N.; Ning, D.; Shi, Z.; Zhou, X.; Wu, L.; Yang, Y.; et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Chang. 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Custodio, M.; Espinoza, C.; Peñaloza, R.; Peralta-Ortiz, T.; Sánchez-Suárez, H.; Ordinola-Zapata, A.; Vieyra-Peña, E. Microbial diversity in intensively farmed lake sediment contaminated by heavy metals and identification of microbial taxa bioindicators of environmental quality. Sci. Rep. 2022, 12, 80. [Google Scholar] [CrossRef]
- Jiao, S.; Yang, Y.; Xu, Y.; Zhang, J.; Lu, Y. Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 2020, 14, 202–216. [Google Scholar] [CrossRef]
- Huo, Y.; Kang, J.-P.; Park, J.-K.; Li, J.; Chen, L.; Yang, D.-C. Rhodanobacter ginsengiterrae sp. nov., an antagonistic bacterium against root rot fungal pathogen Fusarium solani, isolated from ginseng rhizospheric soil. Arch Microbiol. 2018, 200, 1457–1463. [Google Scholar] [CrossRef]
- De Clercq, D.; Van Trappen, S.; Cleenwerck, I.; Ceustermans, A.; Swings, J.; Coosemans, J.; Ryckeboer, J. Rhodanobacter spathiphylli sp. nov., a gammaproteobacterium isolated from the roots of Spathiphyllum plants grown in a compost-amended potting mix. Int. J. Syst. Evol. Microbiol. 2006, 56, 1755–1759. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, L.; Yan, Z.; Schneijderberg, M.; de Roij, M.; Pijnenburg, R.; Zheng, Q.; Franken, C.; Dechesne, A.; Trindade, L.M.; van Velzen, R.; et al. Synthetic bacterial community derived from a desert rhizosphere confers salt stress resilience to tomato in the presence of a soil microbiome. ISME J. 2022, 16, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; He, X.; Sun, J.; Ma, Y. A Degeneration Gradient of Poplar Trees Contributes to the Taxonomic, Functional, and Resistome Diversity of Bacterial Communities in Rhizosphere Soils. IJMS 2021, 22, 3438. [Google Scholar] [CrossRef]
- Jiao, S.; Xu, Y.; Zhang, J.; Hao, X.; Lu, Y. Core Microbiota in Agricultural Soils and Their Potential Associations with Nutrient Cycling. mSystems 2019, 4, e00313–e00318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Zheng, S.; Cao, C.; Li, C. Tillage practices and straw-returning methods affect topsoil bacterial community and organic C under a rice-wheat cropping system in central China. Sci. Rep. 2016, 6, 33155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bever, J.D.; Platt, T.G.; Morton, E.R. Microbial Population and Community Dynamics on Plant Roots and Their Feedbacks on Plant Communities. Annu. Rev. Microbiol. 2012, 66, 265–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Liu, X.; Zhang, M.; Xing, F. Plant Diversity and Fungal Richness Regulate the Changes in Soil Multifunctionality in a Semi-Arid Grassland. Biology 2022, 11, 870. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, F.; Jia, F.-A.; Guan, M.; Jia, Q.-A.; Sun, Y.; Li, Z. Effects of Transplantation and Microhabitat on Rhizosphere Microbial Communities during the Growth of American Ginseng. Agronomy 2023, 13, 1876. https://doi.org/10.3390/agronomy13071876
Chang F, Jia F-A, Guan M, Jia Q-A, Sun Y, Li Z. Effects of Transplantation and Microhabitat on Rhizosphere Microbial Communities during the Growth of American Ginseng. Agronomy. 2023; 13(7):1876. https://doi.org/10.3390/agronomy13071876
Chicago/Turabian StyleChang, Fan, Feng-An Jia, Min Guan, Qing-An Jia, Yan Sun, and Zhi Li. 2023. "Effects of Transplantation and Microhabitat on Rhizosphere Microbial Communities during the Growth of American Ginseng" Agronomy 13, no. 7: 1876. https://doi.org/10.3390/agronomy13071876
APA StyleChang, F., Jia, F.-A., Guan, M., Jia, Q.-A., Sun, Y., & Li, Z. (2023). Effects of Transplantation and Microhabitat on Rhizosphere Microbial Communities during the Growth of American Ginseng. Agronomy, 13(7), 1876. https://doi.org/10.3390/agronomy13071876






