Genome-Wide Identification and Characterization of the PHT1 Gene Family and Its Response to Mycorrhizal Symbiosis in Salvia miltiorrhiza under Phosphate Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. AMF Inoculum Preparation, Plant Materials, and Stress Treatments
2.2. Identification and Phylogenetic Analysis of SmPHT Genes in S. miltiorrhiza
2.3. Gene Structure, Conserved Motif, and Cis-Acting Element Analysis
2.4. RNA Extraction and SmPHT1 Gene Expression
2.5. Analysis of Mycorrhizal Colonization
2.6. Data Statistics and Analysis
3. Results
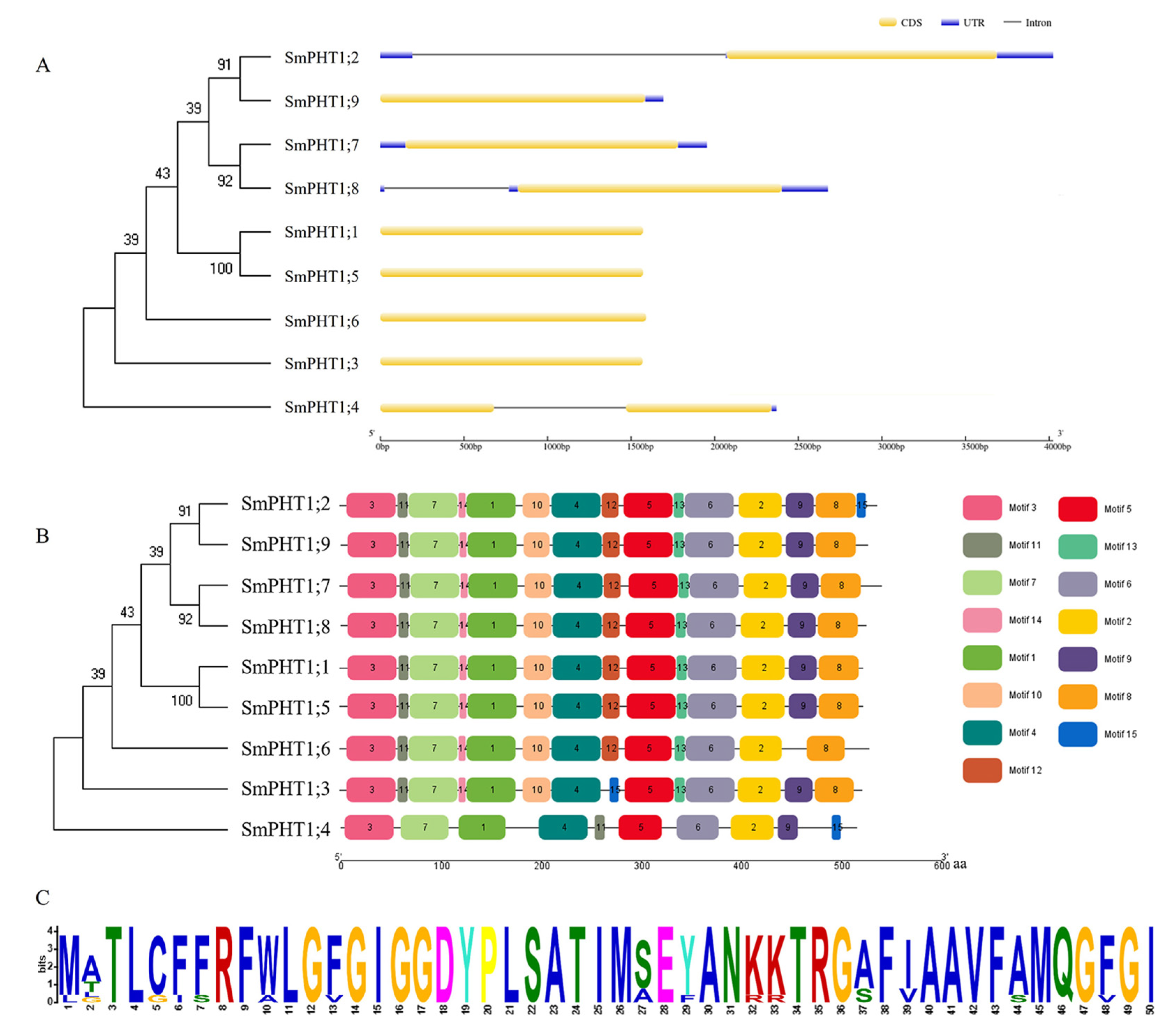
3.1. Identification and Phylogenetic Relationships of SmPHT1
3.2. Gene Structures and Conserved Motifs
3.3. Cis-Acting Element Analysis
3.4. Mycorrhizal Colonization
3.5. Effect of AMF Colonization on the Growth and P uptake of S. miltiorrhiza
3.6. Tissue-Specific Expression Patterns of SmPHT1
3.7. Expression Patterns of SmPHT1 Genes in Response to P and AMF
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, D.; Lv, S.; Jiang, P.; Li, Y. Roles, Regulation, and Agricultural Application of Plant Phosphate Transporters. Front. Plant Sci. 2017, 8, 817. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, L.; Yu, D.; Xu, K.; Zhang, J.; Li, X.; Wang, P.; Chen, G.; Liu, Z.; Peng, C.; et al. Integrative Analysis of the Wheat PHT1 Gene Family Reveals A Novel Member Involved in Arbuscular Mycorrhizal Phosphate Transport and Immunity. Cells 2019, 8, 490. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Jeong, B.R.; Glick, B.R. Contribution of Arbuscular Mycorrhizal Fungi, Phosphate–Solubilizing Bacteria, and Silicon to P Uptake by Plant. Front. Plant Sci. 2021, 12, 699618. [Google Scholar] [CrossRef] [PubMed]
- Karandashov, V.; Bucher, M. Symbiotic Phosphate Transport in Arbuscular Mycorrhizas. Trends Plant Sci. 2005, 10, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Smith, F.A. Roles of Arbuscular Mycorrhizas in Plant Nutrition and Growth: New Paradigms from Cellular to Ecosystem Scales. Annu. Rev. Plant Biol. 2011, 62, 227–250. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Chen, A.; Sun, S.; Xu, G. Complex Regulation of Plant Phosphate Transporters and the Gap between Molecular Mechanisms and Practical Application: What Is Missing? Mol. Plant 2016, 9, 396–416. [Google Scholar] [CrossRef]
- Rui, W.; Ma, J.; Wei, N.; Zhu, X.; Li, Z. Genome-Wide Analysis of the PHT Gene Family and Its Response to Mycorrhizal Symbiosis in Tomatoes under Phosphate Starvation Conditions. Int. J. Mol. Sci. 2023, 24, 10246. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The Unseen Majority: Soil Microbes as Drivers of Plant Diversity and Productivity in Terrestrial Ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Y.; Liao, L.; Xu, J.; Liang, X.; Liu, W. Genome-Wide Identification and Functional Characterization of the Phosphate Transporter Gene Family in Sorghum. Biomolecules 2019, 9, 670. [Google Scholar] [CrossRef]
- Liu, B.; Zhao, S.; Wu, X.; Wang, X.; Nan, Y.; Wang, D.; Chen, Q. Identification and Characterization of Phosphate Transporter Genes in Potato. J. Biotechnol. 2017, 264, 17–28. [Google Scholar] [CrossRef]
- Sun, T.; Li, M.; Shao, Y.; Yu, L.; Ma, F. Comprehensive Genomic Identification and Expression Analysis of the Phosphate Transporter (PHT) Gene Family in Apple. Front. Plant Sci. 2017, 8, 426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Meng, S.; Li, M.; Zhao, Z. Genomic Identification and Expression Analysis of the Phosphate Transporter Gene Family in Poplar. Front. Plant Sci. 2016, 7, 218945. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Deng, M.; Xu, J.; Zhu, X.; Mao, C. Molecular Mechanisms of Phosphate Transport and Signaling in Higher Plants. Semin. Cell Dev. Biol. 2018, 74, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Upadhyay, M.; Srivastava, A.; Abdelrahman, M.; Suprasanna, P.; Tran, L.-S. Cellular and Subcellular Phosphate Transport Machinery in Plants. Int. J. Mol. Sci. 2018, 19, 1914. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, F.; Lu, H.; Liu, Y.; Mao, C. Phosphate Uptake and Transport in Plants: An Elaborate Regulatory System. Plant Cell Physiol. 2021, 62, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Ceasar, S.A.; Baker, A.; Muench, S.P.; Ignacimuthu, S.; Baldwin, S.A. The Conservation of Phosphate-Binding Residues among PHT1 Transporters Suggests That Distinct Transport Affinities Are Unlikely to Result from Differences in the Phosphate-Binding Site. Biochem. Soc. Trans. 2016, 44, 1541–1548. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, A.; David, P.; Arrighi, J.-F.; Chiarenza, S.; Thibaud, M.-C.; Nussaume, L.; Marin, E. Reducing the Genetic Redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 Transporters to Study Phosphate Uptake and Signaling. Plant Physiol. 2015, 167, 1511–1526. [Google Scholar] [CrossRef] [PubMed]
- Paszkowski, U.; Kroken, S.; Roux, C.; Briggs, S.P. Rice Phosphate Transporters Include an Evolutionarily Divergent Gene Specifically Activated in Arbuscular Mycorrhizal Symbiosis. Proc. Natl. Acad. Sci. USA 2002, 99, 13324–13329. [Google Scholar] [CrossRef]
- Wang, M.; Wilde, J.; Baldwin, I.T.; Groten, K. Nicotiana Attenuata ’s Capacity to Interact with Arbuscular Mycorrhiza Alters Its Competitive Ability and Elicits Major Changes in the Leaf Transcriptome. J. Integr. Plant Biol. 2018, 60, 242–261. [Google Scholar] [CrossRef]
- Volpe, V.; Giovannetti, M.; Sun, X.; Fiorilli, V.; Bonfante, P. The Phosphate Transporters LjPT4 and MtPT4 Mediate Early Root Responses to Phosphate Status in Non Mycorrhizal Roots. Plant Cell Environ. 2016, 39, 660–671. [Google Scholar] [CrossRef]
- Chen, A.; Hu, J.; Sun, S.; Xu, G. Conservation and Divergence of Both Phosphate- and Mycorrhiza-regulated Physiological Responses and Expression Patterns of Phosphate Transporters in Solanaceous Species. New Phytol. 2007, 173, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Peters, R.J.; Weirather, J.; Luo, H.; Liao, B.; Zhang, X.; Zhu, Y.; Ji, A.; Zhang, B.; Hu, S.; et al. Full-length Transcriptome Sequences and Splice Variants Obtained by a Combination of Sequencing Platforms Applied to Different Root Tissues of Salvia miltiorrhiza and Tanshinone Biosynthesis. Plant J. 2015, 82, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yuan, L.; Wu, B.; Li, X.; Chen, S.; Lu, S. Genome-Wide Identification and Characterization of Novel Genes Involved in Terpenoid Biosynthesis in Salvia Miltiorrhiza. J. Exp. Bot. 2012, 63, 2809–2823. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, C.; Lu, S. Identification and Characterization of the Cytosine-5 DNA Methyltransferase Gene Family in Salvia miltiorrhiza. PeerJ 2018, 6, e4461. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Huang, F.; Deng, C.; Wang, Y.; Kai, G. Bioactivities, Biosynthesis and Biotechnological Production of Phenolic Acids in Salvia miltiorrhiza. Crit. Rev. Food Sci. Nutr. 2019, 59, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ou, X.; Yang, G.; Xia, Y.; Chen, M.; Guo, L.; Liu, D. Arbuscular Mycorrhizal Fungi Regulate the Growth and Phyto-Active Compound of Salvia Miltiorrhiza Seedlings. Appl. Sci. 2017, 7, 68. [Google Scholar] [CrossRef]
- Bai, Y.; Zhou, Y.; Lei, Q.; Wang, Y.; Pu, G.; Liu, Z.; Chen, X.; Liu, Q. Analysis of the HD-Zip I Transcription Factor Family in Salvia Miltiorrhiza and Functional Research of SmHD-Zip12 in Tanshinone Synthesis. PeerJ 2023, 11, e15510. [Google Scholar] [CrossRef] [PubMed]
- Veneklaas, E.J.; Stevens, J.; Cawthray, G.R.; Turner, S.; Lambers, H. Chickpea and White Lupin Rhizosphere Carboxylates Vary with Soil Properties and Enhance Phosphorus Uptake. Plant Soil 2003, 248, 187–197. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-58829-343-5. [Google Scholar]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A Package of Web Servers for Predicting Subcellular Localization of Proteins in Various Organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Li, P.; Xin, J.; Chen, S.; Zhao, K.; Wu, D.; Fan, Q.; Gao, T.; Chen, F.; Guan, Z. Transcriptome-Wide Survey and Expression Profile Analysis of Putative Chrysanthemum HD-Zip I and II Genes. Genes 2016, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Phillips, J.M.; Hayman, D.S. Improved Procedures for Clearing Roots and Staining Parasitic and Vesicular-Arbuscular Mycorrhizal Fungi for Rapid Assessment of Infection. Trans. Br. Mycol. Soc. 1970, 55, 158-IN18. [Google Scholar] [CrossRef]
- McGONIGLE, T.P.; Miller, M.H.; Evans, D.G.; Fairchild, G.L.; Swan, J.A. A New Method Which Gives an Objective Measure of Colonization of Roots by Vesicular—Arbuscular Mycorrhizal Fungi. New Phytol. 1990, 115, 495–501. [Google Scholar] [CrossRef]
- Fontenot, E.B.; Ditusa, S.F.; Kato, N.; Olivier, D.M.; Dale, R.; Lin, W.; Chiou, T.; Macnaughtan, M.A.; Smith, A.P. Increased Phosphate Transport of Arabidopsis thaliana PHt1;1 by Site-directed Mutagenesis of Tyrosine 312 May Be Attributed to the Disruption of Homomeric Interactions. Plant Cell Environ. 2015, 38, 2012–2022. [Google Scholar] [CrossRef]
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression Analysis Suggests Novel Roles for Members of the Pht1 Family of Phosphate Transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Huang, W.; Liu, F.; Tang, N.; Liu, Y.; Lin, H.; Zhao, B. Functional Analysis of the Novel Mycorrhiza-specific Phosphate Transporter AsPT 1 and PHT 1 Family from Astragalus sinicus during the Arbuscular Mycorrhizal Symbiosis. New Phytol. 2013, 198, 836–852. [Google Scholar] [CrossRef]
- Teng, W.; Zhao, Y.-Y.; Zhao, X.-Q.; He, X.; Ma, W.-Y.; Deng, Y.; Chen, X.-P.; Tong, Y.-P. Genome-Wide Identification, Characterization, and Expression Analysis of PHT1 Phosphate Transporters in Wheat. Front. Plant Sci. 2017, 8, 543. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Zhang, H.; Wang, S.; Ye, X.; Shi, L.; Xu, F.; Ding, G. Molecular Identification of the Phosphate Transporter Family 1 (PHT1) Genes and Their Expression Profiles in Response to Phosphorus Deprivation and Other Abiotic Stresses in Brassica napus. PLoS ONE 2019, 14, e0220374. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Chen, X.; Wang, H.; Liao, D.; Gu, M.; Qu, H.; Sun, S.; Xu, G. Genome-Wide Investigation and Expression Analysis Suggest Diverse Roles and Genetic Redundancy of Pht1 Family Genes in Response to Pi Deficiency in Tomato. BMC Plant Biol 2014, 14, 61. [Google Scholar] [CrossRef] [PubMed]
- Grün, A.; Buchner, P.; Broadley, M.R.; Hawkesford, M.J. Identification and Expression Profiling of Pht1 Phosphate Transporters in Wheat in Controlled Environments and in the Field. Plant Biol. J. 2018, 20, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; He, S.; Li, F.; Li, Z.; Ding, M.; Liu, Q.; Rong, J. Analyses of the Sucrose Synthase Gene Family in Cotton: Structure, Phylogeny and Expression Patterns. BMC Plant Biol. 2012, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Liu, F.; Zhou, B. Genome-Wide Identification and Expression Profile Analysis of the PHT1 Gene Family in Gossypium Hirsutum and Its Two Close Relatives of Subgenome Donor Species. Int. J. Mol. Sci. 2020, 21, 4905. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xiao, L.; Yang, H.; Chen, G.; Zeng, H.; Zhao, H.; Zhu, Y. Genome-Wide Identification, Expression Profiling, and Evolution of Phosphate Transporter Gene Family in Green Algae. Front. Genet. 2020, 11, 590947. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, T.; Wu, X.; Zhao, Z. Molecular Cloning and Functional Analysis of a H+-Dependent Phosphate Transporter Gene from the Ectomycorrhizal Fungus Boletus Edulis in Southwest China. Fungal Biol. 2014, 118, 453–461. [Google Scholar] [CrossRef]
- Liu, P.; Chen, S.; Song, A.; Zhao, S.; Fang, W.; Guan, Z.; Liao, Y.; Jiang, J.; Chen, F. A Putative High Affinity Phosphate Transporter, CmPT1, Enhances Tolerance to Pi Deficiency of Chrysanthemum. BMC Plant Biol. 2014, 14, 18. [Google Scholar] [CrossRef]
- Liu, F.; Chang, X.-J.; Ye, Y.; Xie, W.-B.; Wu, P.; Lian, X.-M. Comprehensive Sequence and Whole-Life-Cycle Expression Profile Analysis of the Phosphate Transporter Gene Family in Rice. Mol. Plant 2011, 4, 1105–1122. [Google Scholar] [CrossRef]
- Cao, D.; Liu, Y.; Ma, L.; Liu, Z.; Li, J.; Wen, B.; Zhang, X.; Yin, P.; Jin, X.; Huang, J. Genome-Wide Identification and Characterization of Phosphate Transporter Gene Family Members in Tea Plants (Camellia sinensis L. O. Kuntze) under Different Selenite Levels. Plant Physiol. Biochem. 2021, 166, 668–676. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. DNA-Binding Domains of Plant-Specific Transcription Factors: Structure, Function, and Evolution. Trends Plant Sci. 2013, 18, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Sirohi, P.; Chauhan, H.; Kumar, R. The Enhanced Phosphorus Use Efficiency in Phosphate-Deficient and Mycorrhiza-Inoculated Barley Seedlings Involves Activation of Different Sets of PHT1 Transporters in Roots. Planta 2021, 254, 38. [Google Scholar] [CrossRef] [PubMed]
- Nussaume, L. Phosphate Import in Plants: Focus on the PHT1 Transporters. Front. Plant Sci. 2011, 2, 83. [Google Scholar] [CrossRef] [PubMed]
- Shukla, V.; Kaur, M.; Aggarwal, S.; Bhati, K.K.; Kaur, J.; Mantri, S.; Pandey, A.K. Tissue Specific Transcript Profiling of Wheat Phosphate Transporter Genes and Its Association with Phosphate Allocation in Grains. Sci. Rep. 2016, 6, 39293. [Google Scholar] [CrossRef] [PubMed]
- An, N.; Huang, J.; Xue, Y.; Liu, P.; Liu, G.; Zhu, S.; Chen, Z. Characterization of Phosphate Transporter Genes and the Function of SgPT1 Involved in Phosphate Uptake in Stylosanthes Guianensis. Plant Physiol. Biochem. 2023, 194, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Versaw, W.K.; Pumplin, N.; Gomez, S.K.; Blaylock, L.A.; Harrison, M.J. Closely Related Members of the Medicago Truncatula PHT1 Phosphate Transporter Gene Family Encode Phosphate Transporters with Distinct Biochemical Activities. J. Biol. Chem. 2008, 283, 24673–24681. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Guo, Y.; Chen, L.; Liang, R.; Gu, M.; Xu, G.; Zhao, J.; Walk, T.; Liao, H. Functional Characterization of 14 Pht1 Family Genes in Yeast and Their Expressions in Response to Nutrient Starvation in Soybean. PLoS ONE 2012, 7, e47726. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhang, H.; Zhang, X.; Chen, H.; Tang, M. Characterization of Six PHT1 Members in Lycium Barbarum and Their Response to Arbuscular Mycorrhiza and Water Stress. Tree Physiol. 2017, 37, 351–366. [Google Scholar] [CrossRef] [PubMed]
- MacLean, A.M.; Bravo, A.; Harrison, M.J. Plant Signaling and Metabolic Pathways Enabling Arbuscular Mycorrhizal Symbiosis. Plant Cell 2017, 29, 2319–2335. [Google Scholar] [CrossRef]
- Raphael, B.; Nicolás, M.; Martina, J.; Daphnée, B.; Daniel, W.; Pierre-Emmanuel, C. The Fine-tuning of Mycorrhizal Pathway in Sorghum Depends on Both Nitrogen−phosphorus Availability and the Identity of the Fungal Partner. Plant Cell Environ. 2022, 45, 3354–3366. [Google Scholar] [CrossRef]
- Breuillin, F.; Schramm, J.; Hajirezaei, M.; Ahkami, A.; Favre, P.; Druege, U.; Hause, B.; Bucher, M.; Kretzschmar, T.; Bossolini, E.; et al. Phosphate Systemically Inhibits Development of Arbuscular Mycorrhiza in Petunia Hybrida and Represses Genes Involved in Mycorrhizal Functioning: Phosphate and Petunia Mycorrhiza Development and Functioning. Plant J. 2010, 64, 1002–1017. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Guo, H.; Zhang, Q.; Guo, H.; Zhang, L.; Zhang, C.; Gou, Z.; Liu, Y.; Wei, J.; Chen, A.; et al. Arbuscular Mycorrhizal Fungi (AMF) Enhanced the Growth, Yield, Fiber Quality and Phosphorus Regulation in Upland Cotton (Gossypium hirsutum L.). Sci. Rep. 2020, 10, 2084. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, R.G.; De Oliveira, V.H.; De Andrade, S.A.L. Arbuscular Mycorrhizal Symbiosis Alters the Expression of PHT1 Phosphate Transporters in Roots and Nodules of P-Starved Soybean Plants. Theor. Exp. Plant Physiol. 2020, 32, 243–253. [Google Scholar] [CrossRef]
- Walder, F.; Brulé, D.; Koegel, S.; Wiemken, A.; Boller, T.; Courty, P. Plant Phosphorus Acquisition in a Common Mycorrhizal Network: Regulation of Phosphate Transporter Genes of the Pht1 Family in Sorghum and Flax. New Phytol. 2015, 205, 1632–1645. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xu, Y.; Jiang, H.; Jiang, C.; Du, Y.; Gong, C.; Wang, W.; Zhu, S.; Han, G.; Cheng, B. Systematic Identification, Evolution and Expression Analysis of the Zea Mays PHT1 Gene Family Reveals Several New Members Involved in Root Colonization by Arbuscular Mycorrhizal Fungi. Int. J. Mol. Sci. 2016, 17, 930. [Google Scholar] [CrossRef]
- Che, X.; Wang, S.; Ren, Y.; Xie, X.; Hu, W.; Chen, H.; Tang, M. A Eucalyptus Pht1 Family Gene EgPT8 Is Essential for Arbuscule Elongation of Rhizophagus irregularis. Microbiol. Spectr. 2022, 10, e01470-22. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Drijber, R.A.; Li, X.; Miller, D.N.; Wienhold, B.J. Arbuscular Mycorrhizal Fungi Differ in Their Ability to Regulate the Expression of Phosphate Transporters in Maize (Zea mays L.). Mycorrhiza 2013, 23, 507–514. [Google Scholar] [CrossRef]
- Yang, S.-Y.; Grønlund, M.; Jakobsen, I.; Grotemeyer, M.S.; Rentsch, D.; Miyao, A.; Hirochika, H.; Kumar, C.S.; Sundaresan, V.; Salamin, N.; et al. Nonredundant Regulation of Rice Arbuscular Mycorrhizal Symbiosis by Two Members of the PHOSPHATE TRANSPORTER1 Gene Family. Plant Cell 2012, 24, 4236–4251. [Google Scholar] [CrossRef]





| Name | Amino Acids | Molecular Weight (Da) | Theoretical pI | Instability Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|
| SmPHT1;1 | 523 | 57,218.7 | 8.32 | 30.35 S | 0.376 | Plasma membrane |
| SmPHT1;2 | 537 | 58,947.83 | 8.86 | 32.44 S | 0.312 | Plasma membrane |
| SmPHT1;3 | 522 | 56,466.87 | 9.01 | 35.33 S | 0.417 | Plasma membrane |
| SmPHT1;4 | 516 | 55,704.84 | 8.83 | 43.73 US | 0.443 | Plasma membrane |
| SmPHT1;5 | 523 | 57,184.76 | 8.64 | 30.96 S | 0.37 | Plasma membrane |
| SmPHT1;6 | 529 | 58,482.94 | 8.67 | 32.11 S | 0.249 | Plasma membrane |
| SmPHT1;7 | 542 | 59,451.05 | 8.98 | 31.81 S | 0.281 | Plasma membrane |
| SmPHT1;8 | 525 | 57,433.07 | 9.15 | 30.96 S | 0.336 | Plasma membrane |
| SmPHT1;9 | 527 | 57,588.11 | 9.07 | 38.79 S | 0.32 | Plasma membrane |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Bai, Y.; Lin, Y.; Liu, H.; Han, F.; Chang, H.; Li, M.; Liu, Q. Genome-Wide Identification and Characterization of the PHT1 Gene Family and Its Response to Mycorrhizal Symbiosis in Salvia miltiorrhiza under Phosphate Stress. Genes 2024, 15, 589. https://doi.org/10.3390/genes15050589
Chen X, Bai Y, Lin Y, Liu H, Han F, Chang H, Li M, Liu Q. Genome-Wide Identification and Characterization of the PHT1 Gene Family and Its Response to Mycorrhizal Symbiosis in Salvia miltiorrhiza under Phosphate Stress. Genes. 2024; 15(5):589. https://doi.org/10.3390/genes15050589
Chicago/Turabian StyleChen, Xue, Yanhong Bai, Yanan Lin, Hongyan Liu, Fengxia Han, Hui Chang, Menglin Li, and Qian Liu. 2024. "Genome-Wide Identification and Characterization of the PHT1 Gene Family and Its Response to Mycorrhizal Symbiosis in Salvia miltiorrhiza under Phosphate Stress" Genes 15, no. 5: 589. https://doi.org/10.3390/genes15050589





