Genotype–Phenotype Correlations in Alport Syndrome—A Single-Center Experience
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Objectives
2.2. Clinical Data
2.3. Genetic Testing
2.4. Statistical Analysis
2.5. Ethics Approval and Consent to Participate
3. Results
3.1. Study Population
3.2. Clinical Features
3.3. Pathological Findings
3.4. Laboratory Findings
3.5. Treatment History
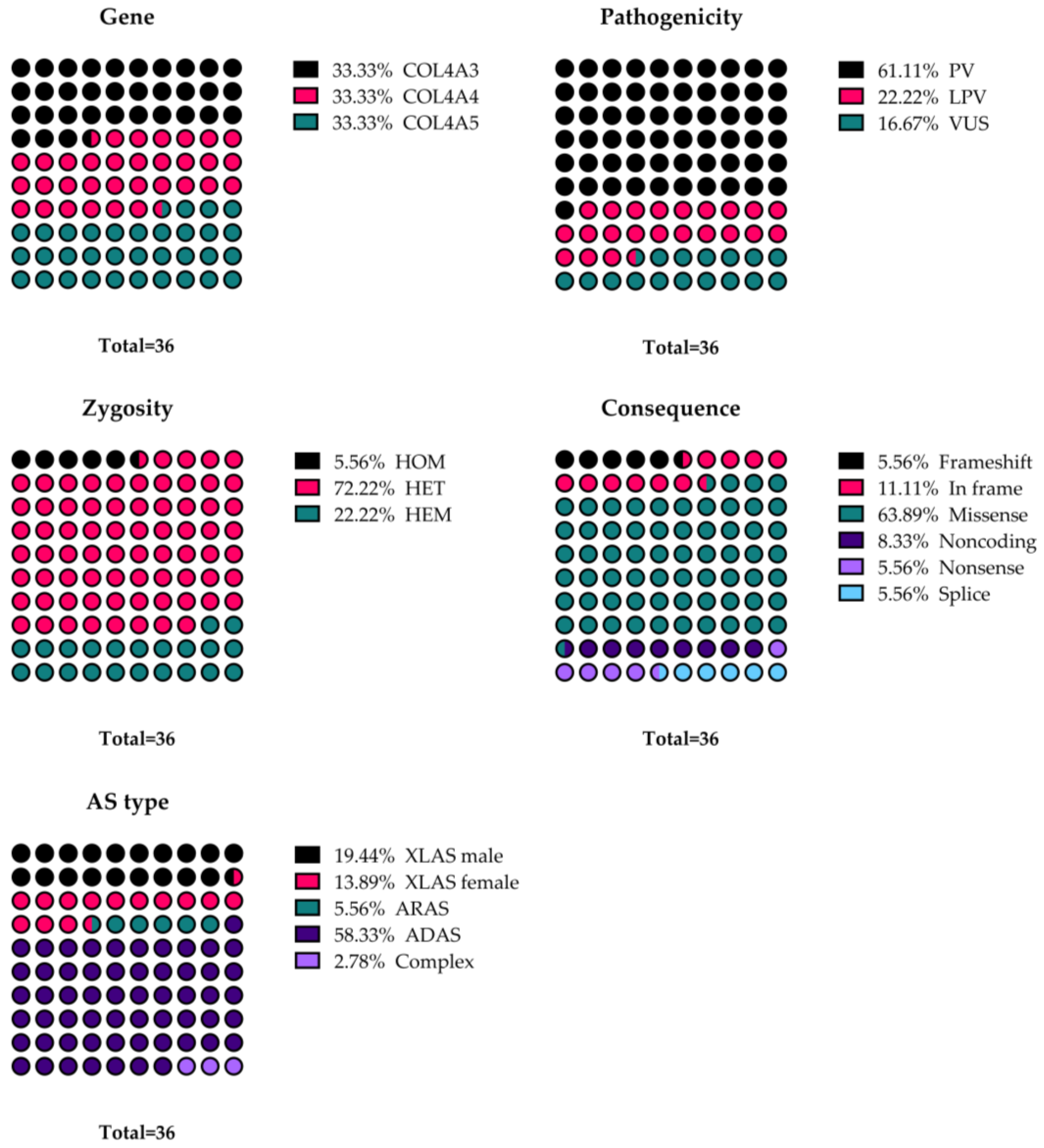
3.6. Genetic Testing
3.7. Renal Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kruegel, J.; Rubel, D.; Gross, O. Alport Syndrome—Insights from Basic and Clinical Research. Nat. Rev. Nephrol. 2012, 9, 170–178. [Google Scholar] [CrossRef]
- Hasstedt, S.J.; Atkin, C.L. X-Linked Inheritance of Alport Syndrome: Family P Revisited. Am. J. Hum. Genet. 1983, 35, 1241–1251. [Google Scholar]
- Pajari, H.; Kääriäinen, H.; Muhonen, T.; Koskimies, O. Alport’s Syndrome in 78 Patients: Epidemiological and Clinical Study. Acta Paediatr. 1996, 85, 1300–1306. [Google Scholar] [CrossRef]
- Gibson, J.; Fieldhouse, R.; Chan, M.M.Y.; Sadeghi-Alavijeh, O.; Burnett, L.; Izzi, V.; Persikov, A.V.; Gale, D.P.; Storey, H.; Savige, J. Prevalence Estimates of Predicted Pathogenic Col4a3-Col4a5 Variants in a Population Sequencing Database and Their Implications for Alport Syndrome. J. Am. Soc. Nephrol. 2021, 32, 2273–2290. [Google Scholar] [CrossRef]
- De Gregorio, V.; Caparali, E.B.; Shojaei, A.; Ricardo, S.; Barua, M. Alport Syndrome: Clinical Spectrum and Therapeutic Advances. Kidney Med. 2023, 5, 100631. [Google Scholar] [CrossRef]
- Savige, J.; Gregory, M.; Gross, O.; Kashtan, C.; Ding, J.; Flinter, F. Expert Guidelines for the Management of Alport Syndrome and Thin Basement Membrane Nephropathy. J. Am. Soc. Nephrol. 2013, 24, 364–375. [Google Scholar] [CrossRef]
- Savige, J.; Lipska-Zietkiewicz, B.S.; Watson, E.; Hertz, J.M.; Deltas, C.; Mari, F.; Hilbert, P.; Plevova, P.; Byers, P.; Cerkauskaite, A.; et al. Guidelines for Genetic Testing and Management of Alport Syndrome. Clin. J. Am. Soc. Nephrol. 2022, 17, 143–154. [Google Scholar] [CrossRef]
- Quinlan, C.; Rheault, M.N. Genetic Basis of Type Iv Collagen Disorders of the Kidney. Clin. J. Am. Soc. Nephrol. 2021, 16, 1101–1109. [Google Scholar] [CrossRef]
- Xie, J.; Wu, X.; Ren, H.; Wang, W.; Wang, Z.; Pan, X.; Hao, X.; Tong, J.; Ma, J.; Ye, Z.; et al. COL4A3 Mutations Cause Focal Segmental Glomerulosclerosis. J. Mol. Cell Biol. 2014, 6, 498–505. [Google Scholar] [CrossRef]
- Papazachariou, L.; Demosthenous, P.; Pieri, M.; Papagregoriou, G.; Savva, I.; Stavrou, C.; Zavros, M.; Athanasiou, Y.; Ioannou, K.; Patsias, C.; et al. Frequency of COL4A3/COL4A4 Mutations amongst Families Segregating Glomerular Microscopic Hematuria and Evidence for Activation of the Unfolded Protein Response. Focal and Segmental Glomerulosclerosis Is a Frequent Development during Ageing. PLoS ONE 2014, 9, e115015. [Google Scholar] [CrossRef]
- Groopman, E.E.; Marasa, M.; Cameron-Christie, S.; Petrovski, S.; Aggarwal, V.S.; Milo-Rasouly, H.; Li, Y.; Zhang, J.; Nestor, J.; Krithivasan, P.; et al. Diagnostic Utility of Exome Sequencing for Kidney Disease. N. Engl. J. Med. 2019, 380, 142–151. [Google Scholar] [CrossRef]
- Storey, H.; Savige, J.; Sivakumar, V.; Abbs, S.; Flinter, F.A. COL4A3/COL4A4 Mutations and Features in Individuals with Autosomal Recessive Alport Syndrome. J. Am. Soc. Nephrol. 2013, 24, 1945–1954. [Google Scholar] [CrossRef]
- Matthaiou, A.; Poulli, T.; Deltas, C. Prevalence of Clinical, Pathological and Molecular Features of Glomerular Basement Membrane Nephropathy Caused by COL4A3 or COL4A4 Mutations: A Systematic Review. Clin. Kidney J. 2020, 13, 1025–1036. [Google Scholar] [CrossRef]
- Bekheirnia, M.R.; Reed, B.; Gregory, M.C.; McFann, K.; Shamshirsaz, A.A.; Masoumi, A.; Schrier, R.W. Genotype-Phenotype Correlation in X-Linked Alport Syndrome. J. Am. Soc. Nephrol. 2010, 21, 876–883. [Google Scholar] [CrossRef]
- Gibson, J.T.; Huang, M.; Shenelli Croos Dabrera, M.; Shukla, K.; Rothe, H.; Hilbert, P.; Deltas, C.; Storey, H.; Lipska-Ziętkiewicz, B.S.; Chan, M.M.Y.; et al. Genotype-Phenotype Correlations for COL4A3-COL4A5 Variants Resulting in Gly Substitutions in Alport Syndrome. Sci. Rep. 2022, 12, 2722. [Google Scholar] [CrossRef]
- Furlano, M.; Martínez, V.; Pybus, M.; Arce, Y.; Crespí, J.; del Prado Venegas, M.; Bullich, G.; Domingo, A.; Ayasreh, N.; Benito, S.; et al. Clinical and Genetic Features of Autosomal Dominant Alport Syndrome: A Cohort Study. Am. J. Kidney Dis. 2021, 78, 560–570.e1. [Google Scholar] [CrossRef]
- Inker, L.A.; Eneanya, N.D.; Coresh, J.; Tighiouart, H.; Wang, D.; Sang, Y.; Crews, D.C.; Doria, A.; Estrella, M.M.; Froissart, M.; et al. New Creatinine- and Cystatin C–Based Equations to Estimate GFR without Race. N. Engl. J. Med. 2021, 385, 1737–1749. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The Human Genomic Variant Search Engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Jais, J.P.; Knebelmann, B.; Giatras, I.; De Marchi, M.; Rizzoni, G.; Renieri, A.; Weber, M.; Gross, O.; Netzer, K.-O.; Flinter, F.; et al. X-Linked Alport Syndrome. J. Am. Soc. Nephrol. 2000, 11, 649–657. [Google Scholar] [CrossRef]
- Gross, O.; Netzer, K.O.; Lambrecht, R.; Seibold, S.; Weber, M. Meta-analysis of Genotype–Phenotype Correlation in X-linked Alport Syndrome: Impact on Clinical Counselling. Nephrol. Dial. Transplant. 2002, 17, 1218–1227. [Google Scholar] [CrossRef]
- Persikov, A.V.; Pillitteri, R.J.; Amin, P.; Schwarze, U.; Byers, P.H.; Brodsky, B. Stability Related Bias in Residues Replacing Glycines within the Collagen Triple Helix (Gly-Xaa-Yaa) in Inherited Connective Tissue Disorders. Hum. Mutat. 2004, 24, 330–337. [Google Scholar] [CrossRef]
- García-Aznar, J.M.; De la Higuera, L.; Besada Cerecedo, L.; Gandiaga, N.P.; Vega, A.I.; Fernández-Fresnedo, G.; González-Lamuño, D. New Insights into Renal Failure in a Cohort of 317 Patients with Autosomal Dominant Forms of Alport Syndrome: Report of Two Novel Heterozygous Mutations in COL4A3. J. Clin. Med. 2022, 11, 4883. [Google Scholar] [CrossRef]
- Zhang, Y.; Ding, J.; Zhang, H.; Yao, Y.; Xiao, H.; Wang, S.; Wang, F. Effect of Heterozygous Pathogenic COL4A3 or COL4A4 Variants on Patients with X-Linked Alport Syndrome. Mol. Genet. Genom. Med. 2019, 7, e647. [Google Scholar] [CrossRef]
- Mencarelli, M.A.; Heidet, L.; Storey, H.; Van Geel, M.; Knebelmann, B.; Fallerini, C.; Miglietti, N.; Antonucci, M.F.; Cetta, F.; Sayer, J.A.; et al. Evidence of Digenic Inheritance in Alport Syndrome. J. Med. Genet. 2015, 52, 163–174. [Google Scholar] [CrossRef]
- Frascà, G.M.; Onetti-Muda, A.; Mari, F.; Longo, I.; Scala, E.; Pescucci, C.; Roccatello, D.; Alpa, M.; Coppo, R.; Li Volti, G.; et al. Thin Glomerular Basement Membrane Disease: Clinical Significance of a Morphological Diagnosis—A Collaborative Study of the Italian Renal Immunopathology Group. Nephrol. Dial. Transplant. 2005, 20, 545–551. [Google Scholar] [CrossRef]
- Chiereghin, C.; Robusto, M.; Mastrangelo, A.; Castorina, P.; Montini, G.; Giani, M.; Duga, S.; Asselta, R.; Soldà, G. Alport Syndrome Cold Cases: Missing Mutations Identified by Exome Sequencing and Functional Analysis. PLoS ONE 2017, 12, e0178630. [Google Scholar] [CrossRef]
- Kamiyoshi, N.; Nozu, K.; Fu, X.J.; Morisada, N.; Nozu, Y.; Ye, M.J.; Imafuku, A.; Miura, K.; Yamamura, T.; Minamikawa, S.; et al. Genetic, Clinical, and Pathologic Backgrounds of Patients with Autosomal Dominant Alport Syndrome. Clin. J. Am. Soc. Nephrol. 2016, 11, 1441–1449. [Google Scholar] [CrossRef]
- Morinière, V.; Dahan, K.; Hilbert, P.; Lison, M.; Lebbah, S.; Topa, A.; Bole-Feysot, C.; Pruvost, S.; Nitschke, P.; Plaisier, E.; et al. Improving Mutation Screening in Familial Hematuric Nephropathies through next Generation Sequencing. J. Am. Soc. Nephrol. 2014, 25, 2740–2751. [Google Scholar] [CrossRef]
- Weber, S.; Strasser, K.; Rath, S.; Kittke, A.; Beicht, S.; Alberer, M.; Lange-Sperandio, B.; Hoyer, P.F.; Benz, M.R.; Ponsel, S.; et al. Identification of 47 Novel Mutations in Patients with Alport Syndrome and Thin Basement Membrane Nephropathy. Pediatr. Nephrol. 2016, 31, 941–955. [Google Scholar] [CrossRef]
- Savige, J.; Storey, H.; Cheong, H.I.; Kang, H.G.; Park, E.; Hilbert, P.; Persikov, A.; Torres-Fernandez, C.; Ars, E.; Torra, R.; et al. X-Linked and Autosomal Recessive Alport Syndrome: Pathogenic Variant Features and Further Genotype-Phenotype Correlations. PLoS ONE 2016, 11, e0161802. [Google Scholar] [CrossRef]
- Macheroux, E.P.; Braunisch, M.C.; Pucci Pegler, S.; Satanovskij, R.; Riedhammer, K.M.; Günthner, R.; Gross, O.; Nagel, M.; Renders, L.; Hoefele, J. The Hypomorphic Variant p.(Gly624Asp) in COL4A5 as a Possible Cause for an Unexpected Severe Phenotype in a Family With X-Linked Alport Syndrome. Front. Pediatr. 2019, 7, 485. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving Access to Variant Interpretations and Supporting Evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Fokkema, I.F.A.C.; Taschner, P.E.M.; Schaafsma, G.C.P.; Celli, J.; Laros, J.F.J.; den Dunnen, J.T. LOVD v.2.0: The next Generation in Gene Variant Databases. Hum. Mutat. 2011, 32, 557–563. [Google Scholar] [CrossRef]
- Kashtan, C.E.; Gross, O. Clinical Practice Recommendations for the Diagnosis and Management of Alport Syndrome in Children, Adolescents, and Young Adults-an Update for 2020. Pediatr. Nephrol. 2021, 36, 711–719. [Google Scholar] [CrossRef]
- Malone, A.F.; Phelan, P.J.; Hall, G.; Cetincelik, U.; Homstad, A.; Alonso, A.S.; Jiang, R.; Lindsey, T.B.; Wu, G.; Sparks, M.A.; et al. Rare Hereditary COL4A3/COL4A4 Variants May Be Mistaken for Familial Focal Segmental Glomerulosclerosis. Kidney Int. 2014, 86, 1253–1259. [Google Scholar] [CrossRef]
- Gast, C.; Pengelly, R.J.; Lyon, M.; Bunyan, D.J.; Seaby, E.G.; Graham, N.; Venkat-Raman, G.; Ennis, S. Collagen (COL4A) Mutations Are the Most Frequent Mutations Underlying Adult Focal Segmental Glomerulosclerosis. Nephrol. Dial. Transplant. 2016, 31, 961–970. [Google Scholar] [CrossRef]
- Fallerini, C.; Dosa, L.; Tita, R.; Del Prete, D.; Feriozzi, S.; Gai, G.; Clementi, M.; La Manna, A.; Miglietti, N.; Mancini, R.; et al. Unbiased next Generation Sequencing Analysis Confirms the Existence of Autosomal Dominant Alport Syndrome in a Relevant Fraction of Cases. Clin. Genet. 2014, 86, 252–257. [Google Scholar] [CrossRef]
- Savige, J.; Colville, D.; Rheault, M.; Gear, S.; Lennon, R.; Lagas, S.; Finlay, M.; Flinter, F. Alport Syndrome in Women and Girls. Clin. J. Am. Soc. Nephrol. 2016, 11, 1713–1720. [Google Scholar] [CrossRef]
- Yamamura, T.; Horinouchi, T.; Nagano, C.; Omori, T.; Sakakibara, N.; Aoto, Y.; Ishiko, S.; Nakanishi, K.; Shima, Y.; Nagase, H.; et al. Genotype-Phenotype Correlations Influence the Response to Angiotensin-Targeting Drugs in Japanese Patients with Male X-Linked Alport Syndrome. Kidney Int. 2020, 98, 1605–1614. [Google Scholar] [CrossRef]
- Ozdemir, G.; Gulhan, B.; Atayar, E.; Saygılı, S.; Soylemezoglu, O.; Ozcakar, Z.B.; Eroglu, F.K.; Candan, C.; Demir, B.K.; Soylu, A.; et al. COL4A3 Mutation Is an Independent Risk Factor for Poor Prognosis in Children with Alport Syndrome. Pediatr. Nephrol. 2020, 35, 1941–1952. [Google Scholar] [CrossRef] [PubMed]
- Jais, J.P.; Knebelmann, B.; Giatras, I.; De Marchi, M.; Rizzoni, G.; Renieri, A.; Weber, M.; Gross, O.; Netzer, K.O.; Flinter, F.; et al. X-Linked Alport Syndrome: Natural History and Genotype-Phenotype Correlations in Girls and Women Belonging to 195 Families: A “European Community Alport Syndrome Concerted Action” Study. J. Am. Soc. Nephrol. 2003, 14, 2603–2610. [Google Scholar] [CrossRef] [PubMed]
- Boeckhaus, J.; Strenzke, N.; Storz, C.; Gross, O.; on behalf of the GPN Study Group; EARLY PRO-TECT Alport Investigators. Characterization of Sensorineural Hearing Loss in Children with Alport Syndrome. Life 2020, 10, 360. [Google Scholar] [CrossRef] [PubMed]
- Kashtan, C.E. Animal Models of Alport Syndrome. Nephrol. Dial. Transplant. 2002, 17, 1359–1362. [Google Scholar] [CrossRef]
- Zehnder, A.F.; Adams, J.C.; Santi, P.A.; Kristiansen, A.G.; Wacharasindhu, C.; Mann, S.; Kalluri, R.; Gregory, M.C.; Kashtan, C.E.; Merchant, S.N. Distribution of Type IV Collagen in the Cochlea in Alport Syndrome. Arch. Otolaryngol. Head Neck Surg. 2005, 131, 1007–1013. [Google Scholar] [CrossRef]


| Laboratory Findings at Referral | |||
|---|---|---|---|
| Including Patients Presenting with ESKD | Excluding Patients Presenting with ESKD | ||
| Serum Creatinine, mg/dL | |||
| Mean ± SD | 3.83 ± 3.51 | Mean ± SD | 1.56 ± 0.87 |
| No./total (%) | 36/36 (100) | No./total (%) | 21/21 (100) |
| eGFR, mL/min per 1.73 m2 | |||
| Mean ± SD | 46.2 ± 39.01 | Mean ± SD | 65.95 ± 35.28 |
| No./total (%) | 36/36 (100) | No./total (%) | 21/21 (100) |
| Proteinuria, g/day | |||
| Mean ± SD | 2.04 ± 1.53 | Mean ± SD | 1.77 ± 1.44 |
| No./total (%) | 15/36 (41.7) | No./total (%) | 12/21 (57.1) |
| Hematuria, no./total (%) | |||
| Hematuria | 20/26 (76.9) | Hematuria | 15/20 (75) |
| No./total (%) | 26/36 (72.2) | No./total (%) | 20/21 (95.2) |
| Patient Number | Variant Characteristics | Type of AS | Previously Described | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Position | Nucleotide Change | Amino Acid Change | Zygosity | Pathogenicity | Consequence | Inheritance | |||
| 33 | COL4A3 | Exon 26 | c.1814G>T | p.(Gly605Val) | HET | LPV | Missense | AD, AR | ADAS | No * |
| 34 | COL4A3 | Exon 26 | c.1855G>A | p.(Gly619Arg) | HET | PV | Missense | AD, AR | ADAS | Yes [23] |
| 15 | COL4A3 | Exon 32 | c.2549G>A | p.(Gly850Glu) | HET | LPV | Missense | AD, AR | ADAS | Yes [24] |
| 29 | COL4A3 | Intron 14 | c.2746+1G>T | p.? | HET | PV | Noncoding | AD, AR | ADAS | Yes [25] |
| 8, 25 | COL4A3 | Exon 38 | c.3321_3329del | p.(Ser1108_Gly1110del) | HET | LPV | In frame | AD, AR | ADAS | Yes [26] |
| 19 | COL4A3 | Exon 41 | c.3546_3548dup | p.(Gly1183dup) | HET | VUS | In frame | AD, AR | ADAS | No * |
| 1 | COL4A3 | Exon 42 | c.3602G>A | p.(Gly1201Asp) | HOM | LPV | Missense | AD, AR | ARAS | No * |
| 26 | COL4A3 | Exon 44 | c.3925C>T | p.(Pro1309Ser) | HET | VUS | Missense | AD, AR | ADAS | No * |
| 6 | COL4A3 | Exon 1 | c.40_63del | p.(Leu14_Leu21del) | HET | PV | In frame | AD, AR | ADAS | Yes [27] |
| 10 | COL4A3 | Exon 48 | c.4348C>T | p.(Arg1450*) | HET | PV | Nonsense | AD, AR | ADAS | Yes [28] |
| 2 | COL4A3 | Exon 51 | c.4825C>T | p.(Arg1609*) | HOM | PV | Nonsense | AD, AR | ARAS | Yes [28] |
| 22, 32 | COL4A4 | Exon 20 | c.1321_1369+3del | 52bp-Deletion | HET | PV | Splice junction loss | AD, AR | ADAS | Yes [29] |
| 28 | COL4A4 | Exon 24 | c.1716del | p.(Pro573Leufs*80) | HET | PV | Frameshift | AD, AR | ADAS | Yes [29] |
| 3 | COL4A4 | Exon 27 | c.2159C>T | p.(Pro720Leu) | HET | VUS | Missense | AD, AR | ADAS | No * |
| 17, 20, 30, 31 | COL4A4 | Exon 31 | c.2734G>C | p.(Gly912Arg) | HET | LPV/PV ** | Missense | AD, AR | ADAS | No * |
| 36 | COL4A4 | Exon 41 | c.3961del | p.(Asp1321Metfs*67) | HET | PV | Frameshift | Complex inheritance | Complex | Yes [29] |
| 23, 27 | COL4A4 | Exon 48 | c.5045G>A | p.(Arg1682Gln) | HET | VUS | Missense | AD, AR | ADAS | Yes [30] |
| 35 | COL4A4 | Intron 14 | c.871-3A>G | p.? | HET | VUS | Noncoding | AD, AR | ADAS | No * |
| 9 | COL4A5 | Exon 20 | c.1226G>A | p.(Gly409Asp) | HEM | PV | Missense | X-linked | X-linked male | Yes [31] |
| 14, 16 | COL4A5 | Exon 25 | c.1871G>A | p.(Gly624Asp) | HEM | PV | Missense | X-linked | X-linked male | Yes [32] |
| 24 | COL4A5 | Exon 31 | c.2605G>A | p.(Gly869Arg) | HEM | PV | Missense | X-linked | X-linked male | Yes [31] |
| 4, 5, 7, 11, 12, 13 | COL4A5 | Exon 41 | c.3721G>T | p.(Gly1241Cys) | HET/HEM | PV | Missense | X-linked | X-linked male/female | Yes [14] |
| 18 | COL4A5 | Exon 11 | c.637G>C | p.(Gly213Arg) | HET | LPV | Missense | X-linked | X-linked female | Yes [31] |
| 21 | COL4A5 | Intron 12 | c.688-1G>A | p.? | HEM | LPV | Noncoding | X-linked | X-linked male | No * |
| Characteristics | Type of Collagen α Chain | p Value | ||
|---|---|---|---|---|
| COL4A3 | COL4A4 | COL4A5 | ||
| Sex, No. (%) | ||||
| Male | 4/36 (11.1) | 7/36 (19.4) | 8/36 (22.2) | 0.50 |
| Female | 8/36 (22.2) | 5/36 (13.8) | 4/36 (11.1) | 0.46 |
| Laboratory findings | ||||
| Serum creatinine, mg/dL | 4.12 ± 3.34 | 3.75 ± 3.99 | 3.78 ± 3.46 | 0.82 |
| eGFR, mL/min per 1.73 m2 | 40.42 ± 41.36 | 48.33 ± 33.14 | 50.18 ± 44.94 | 0.69 |
| Proteinuria, g/day | 2.2 ± 1.64 | 2.12 ± 1.69 | 1.28 ± 1.01 | 0.77 |
| Hematuria, no./total (%) | 6/7 (85.7) | 8/10 (80) | 6/9 (66.7) | 0.64 |
| ESKD, no./total (%) | ||||
| ESKD at diagnosis | 7/12 (58.3) | 3/12 (25) | 5/12 (41.6) | 0.25 |
| Progression to ESKD | 1/12 (8.3) | 1/12 (8.3) | 1/12 (8.3) | 1 |
| Kidney transplant | 6/12 (50) | 1/12 (8.3) | 4/12 (33.3) | 0.08 |
| Age, median (IQR), y. | ||||
| ESKD | 25 (20–35) | 41 (27.25–53.25) | 24 (21–26.5) | 0.51 |
| Kidney transplant | 27 (22.75–38) | - | 25.5 (23.25–39.75) | 0.38 |
| Mean kidney survival (95% CI), y. | Type of collagen α chain | p value | |||||||||
| COL4A3 | COL4A4 | COL4A5 | Overall | 0.12 | |||||||
| 33.06 (22.79–39.34) | 46.83 (39.89–53.76) | 38.79 (29.79–47.79) | 40.55 (35.55–45.55) | ||||||||
| Inheritance | 0.891 | ||||||||||
| X-linked | Autosomal | Digenic/Complex | Overall | ||||||||
| 38.79 (29.79–47.98) | 41.15 (35.08–47.21) | 40 (40–40) | 40.55 (35.55–45.55) | ||||||||
| Zygosity | 0.054 | ||||||||||
| Homozygous | Heterozygous | Hemizygous | Overall | ||||||||
| 30 (20.2–39.8) | 44.07 (38.55–49.6) | 31 (22.42–39.57) | 40.55 (35.55–45.55) | ||||||||
| Classification | 0.81 | ||||||||||
| Pathogenic | Likely Pathogenic | VUS | Overall | ||||||||
| 38.51 (32.32–44.72) | 40.27 (29.63–50.91) | 43.66 (33.80–53.52) | 40.55 (35.55–45.55) | ||||||||
| Multiple mutations involving COL4 genes | 0.23 | ||||||||||
| Yes | No | Overall | |||||||||
| 36.33 (26.37–46.29) | 41.67 (35.99–47.34) | 40.55 (35.55–45.55) | |||||||||
| Coding impact | 0.023 * | ||||||||||
| Missense | Another type | Overall | |||||||||
| 44.8 (38.87–50.72) | 32.98 (26.05–39.91) | 40.55 (35.55–45.55) | |||||||||
| Type of AS | 0.29 | ||||||||||
| XLAS males | XLAS females | ARAS | ADAS | Complex | |||||||
| 32.16 (22.47–41.86) | 45.75 (33.44–58.05) | 30 (20.2–39.8) | 42.48 (36.08–48.88) | 40 (40–40) | |||||||
| Mean kidney survival (95% CI), y. | Substituted residue (all missense variants) | p value | ||
| Glycine | Another amino acid | Overall | 0.41 | |
| 42.05 (35.49–48.62) | 51.75 (46.23–57.26) | 44.8 (38.87–50.72) | ||
| Substituting residue (glycine substitutions) | 0.92 | |||
| Destabilizing residues (Arg/Val/Glu/Asp/Trp) | Non-destabilizing residues (Ala/Cys/Ser) | Overall | ||
| 42.72 (35.26–50.18) | 39.66 (25.88–53.45) | 42.05 (35.49–48.62) | ||
| Molecular location (all missense variants) | 0.55 | |||
| Exon 1–20 | Exon 21-carboxi terminus | Overall | ||
| 31.5 (16.94–46.05) | 45.53 (39.51–51.51) | 44.8 (38.87–50.72) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lujinschi, Ș.N.; Sorohan, B.M.; Obrișcă, B.; Vrabie, A.; Lupușoru, G.; Achim, C.; Andronesi, A.G.; Covic, A.; Ismail, G. Genotype–Phenotype Correlations in Alport Syndrome—A Single-Center Experience. Genes 2024, 15, 593. https://doi.org/10.3390/genes15050593
Lujinschi ȘN, Sorohan BM, Obrișcă B, Vrabie A, Lupușoru G, Achim C, Andronesi AG, Covic A, Ismail G. Genotype–Phenotype Correlations in Alport Syndrome—A Single-Center Experience. Genes. 2024; 15(5):593. https://doi.org/10.3390/genes15050593
Chicago/Turabian StyleLujinschi, Ștefan Nicolaie, Bogdan Marian Sorohan, Bogdan Obrișcă, Alexandra Vrabie, Gabriela Lupușoru, Camelia Achim, Andreea Gabriella Andronesi, Andreea Covic, and Gener Ismail. 2024. "Genotype–Phenotype Correlations in Alport Syndrome—A Single-Center Experience" Genes 15, no. 5: 593. https://doi.org/10.3390/genes15050593






