Autocatalytic Selection as a Driver for the Origin of Life
Abstract
:1. Introduction
2. Guidelines
2.1. Guideline 1: Specificity Increases over Time
- More specifically, it implies that a primitive genetic code (that is, a relationship between specific nucleotide groups and specific amino acids [41,42,43,44]) must have emerged before the existence of genes and thus that the genetic code was determined largely by chemistry, well before the requirement for genes.
- In the early stages, we should not expect to find pathways that exist in order to produce individual compounds (for example, a pathway leading specifically to glucose or ribose). Rather, we expect nonspecific reaction types that generate a common pool of equilibrating metabolites (e.g., a set of reactions that produce a collection of sugars in equilibrium). Any pathway that requires a specific reaction or product is inherently unlikely.
2.2. Guideline 2: Reactions That Resemble Current Biochemical Reactions Are Inherently More Likely
2.3. Guideline 3: No Teleology
2.4. Guideline 4: Autocatalytic Selection
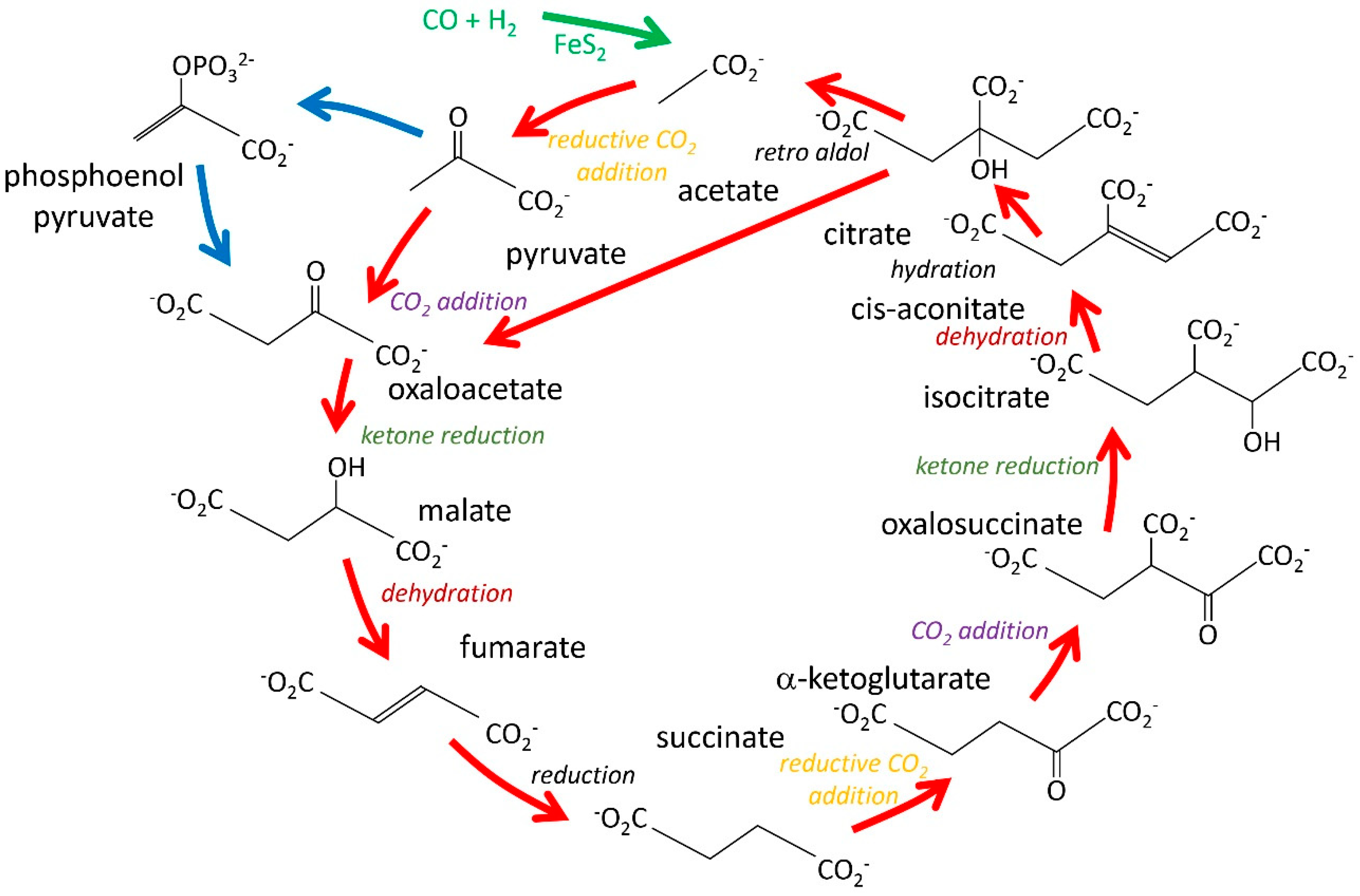
3. Proposals for Autocatalytic Origins of Key Metabolites
3.1. Phosphosugars and the Pentose Phosphate Pathway
3.2. The Autocatalytic Origins of Lipid Membranes
3.3. The Autocatalytic Origins of Amino Acids
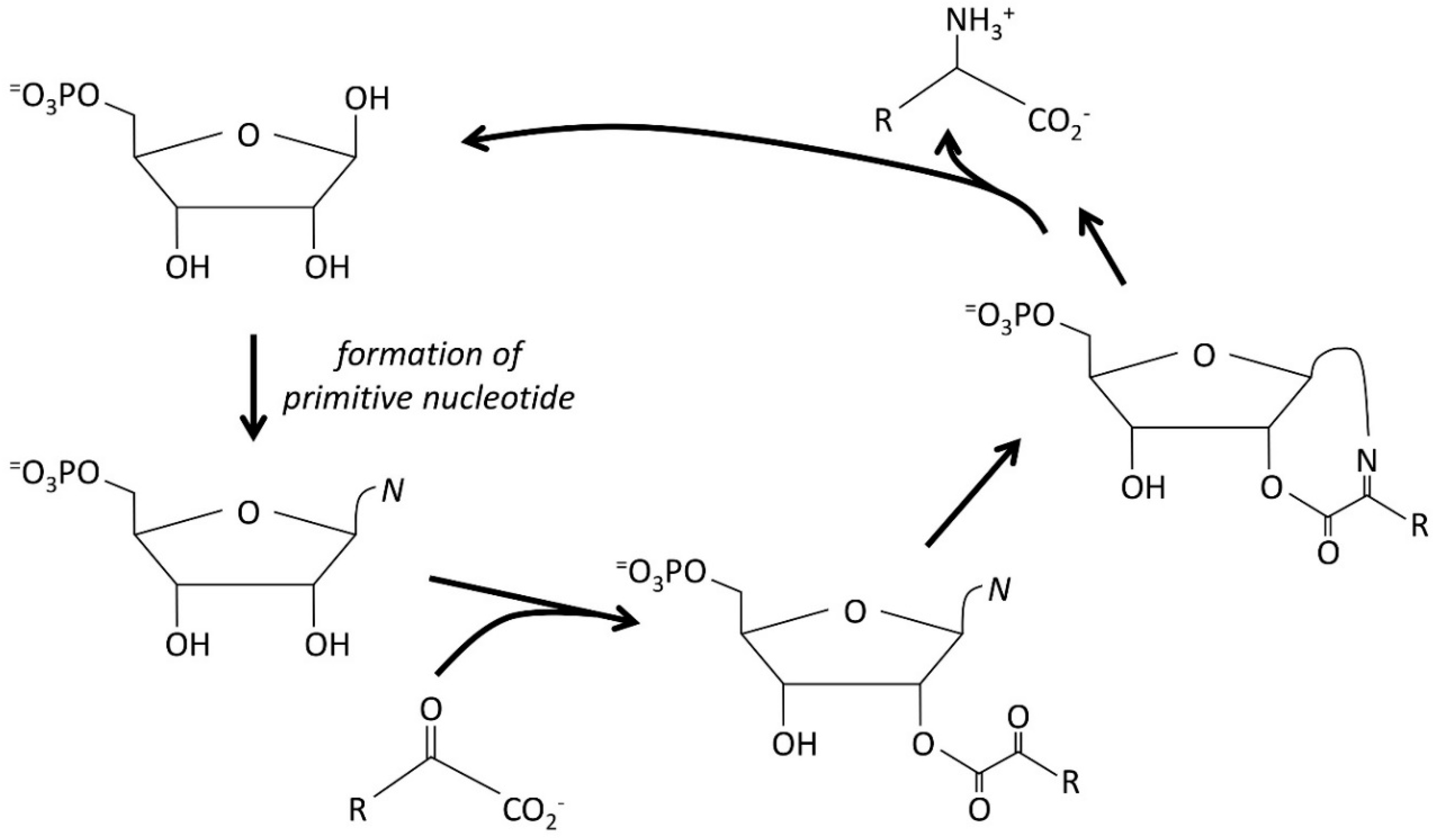
3.4. The Autocatalytic Origins of Nucleic Acids
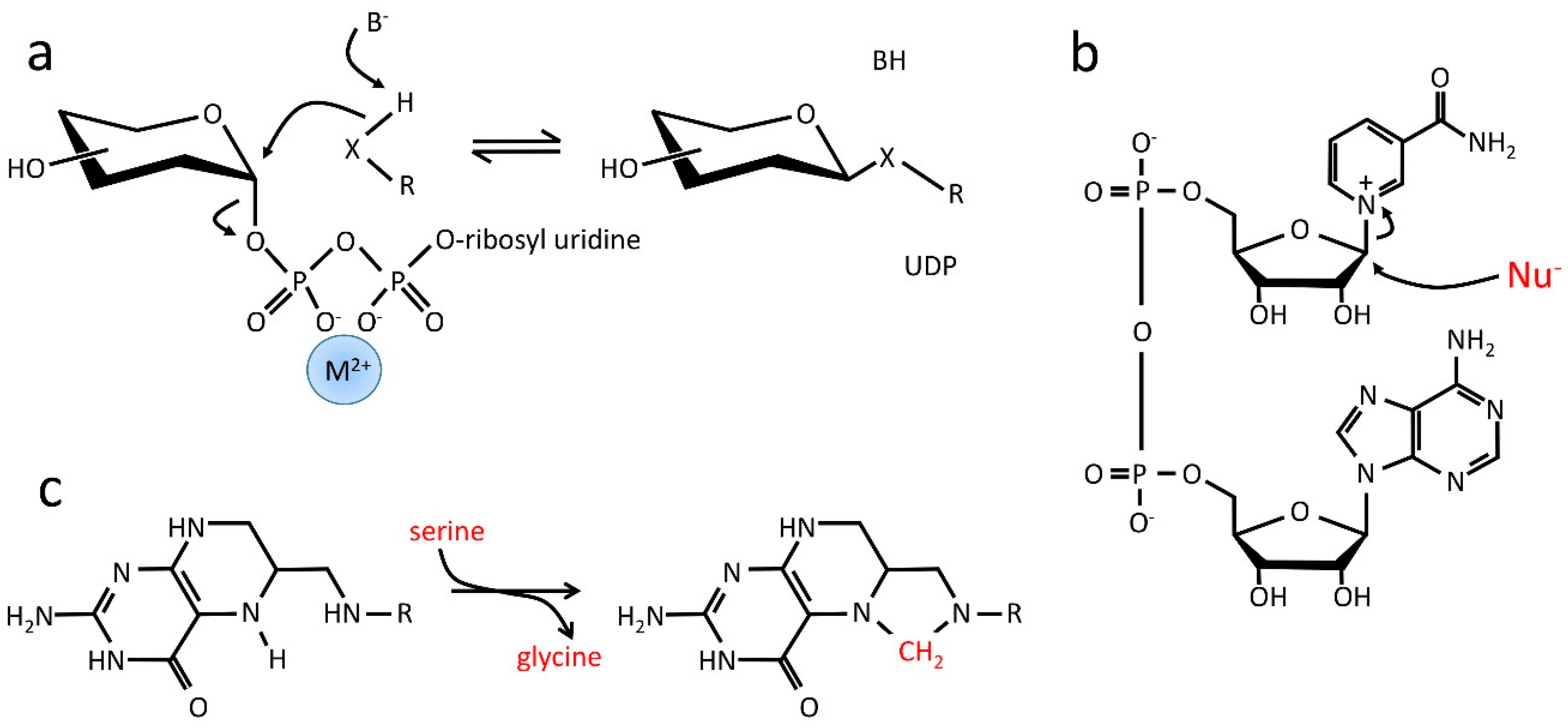
- The vast majority of current reactions that involve molecules with nitrogenous bases do not actually involve the base in the reaction at all. For example, in the reactions in which ATP or GTP is a substrate for phosphoryl transfer, the base is not involved—the entire phosphoribosyl nucleotide is an elaborate leaving group to facilitate phosphoryl transfer, with the remaining diphosphate coordinated to Mg2+ to make it a better leaving group. For reactions involving NAD(P) or FAD in redox reactions, again, the base is not involved in the reaction. In all these cases, the base is used as a “handle” rather than a reactant.
- UDP-glucose acts as a carrier for glucose (Figure 4a); the Mg2+:diphosphoribosyl nucleotide is simply a leaving group [81]. In a similar way, NAD is a substrate for reactions involving poly(ADP-ribosyl) transferases (PARPs), where the nicotinamide ring is a leaving group facilitating the transfer of phosphoadenosylribose to a range of nucleophiles (Figure 4b) [82]. Significantly, in both these reactions, the product is a polymer; the function of the nucleotide diphosphate is to “deliver” a new monomer.
- In folic acid derivatives such as tetrahydrofolate (Figure 4c), the nitrogens act as ligands to bind and release a carbon atom and therefore function as carriers for 1C moieties in a range of oxidation states; they are able to act both as nucleophiles and as leaving groups (i.e., as nucleophilic catalysts).
- There are many nitrogenous compounds, such as NAD(P), FAD and molybdopterins, where the nitrogen-containing ring acts as a cofactor for redox reactions (equivalently, it is a carrier for hydride ions) [83].
3.5. The Autocatalytic Origins of Proteins
4. Discussion
Funding
Data Availability Statement
Conflicts of Interest
References
- Smith, E.; Morowitz, H.J. The Origin and Nature of Life on Earth; Cambridge University Press: Cambridge, UK, 2016. [Google Scholar]
- Lane, N.; Xavier, J.C. To unravel the origin of life, treat findings as pieces of a bigger puzzle. Nature 2024, 626, 948–951. [Google Scholar] [CrossRef]
- Brunk, C.F.; Marshall, C.R. ‘Whole organism’, systems biology, and top-down criteria for evaluating scenarios for the origin of life. Life 2021, 11, 690. [Google Scholar] [CrossRef] [PubMed]
- Wächtershäuser, G. Evolution of the first metabolic cycles. Proc. Natl. Acad. Sci. USA 1990, 87, 200–204. [Google Scholar] [CrossRef]
- Schlesinger, G.; Miller, S.L. Prebiotic synthesis in atmospheres containing CH4, CO, and CO2. 1. Amino acids. J. Mol. Evol. 1983, 19, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Pascal, R.; Pross, A.; Sutherland, J.D. Towards an evolutionary theory of the origin of life based on kinetics and thermodynamics. Open Biol. 2013, 3, 130156. [Google Scholar] [CrossRef] [PubMed]
- Nicolis, G.; Prigogine, I. Self-Organization in Nonequilibrium Systems: From Dissipative Structures to Order through Fluctuations; Wiley: New York, NY, USA, 1977. [Google Scholar]
- Wächtershäuser, G. Before enzymes and templates: Theory of surface metabolism. Microbiol. Rev. 1988, 52, 452–484. [Google Scholar] [CrossRef] [PubMed]
- Benner, S.A.; Ricardo, A.; Carrigan, M.A. Is there a common chemical model for life in the universe? Curr. Opin. Chem. Biol. 2004, 8, 672–689. [Google Scholar] [CrossRef] [PubMed]
- Camprubí, E.; De Leeuw, J.W.; House, C.H.; Raulin, F.; Russell, M.J.; Spang, A.; Tirumalai, M.R.; Westall, F. The emergence of life. Space Sci. Rev. 2019, 215, 56. [Google Scholar] [CrossRef]
- Branscomb, E.; Russell, M.J. Frankenstein or a submarine alkaline vent: Who is responsible for abiogenesis? Part 2: As life is now, so it must have been in the beginning. Bioessays 2018, 40, 1700182. [Google Scholar] [CrossRef]
- Martin, W.F.; Sousa, F.L.; Lane, N. Energy at life’s origin. Science 2014, 344, 1092–1093. [Google Scholar] [CrossRef]
- Cody, G.D.; Boctor, N.Z.; Filley, T.R.; Hazen, R.M.; Scott, J.H.; Sharma, A.; Yoder, H.S. Primordial carbonylated iron-sulfur compounds and the synthesis of pyruvate. Science 2000, 289, 1337–1340. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.A.; Rammu, H.; Liu, F.; Halpern, A.; Palmeira, R.N.; Lane, N. Life as a guide to its own origins. Annu. Rev. Ecol. Evol. Syst. 2023, 54, 327–350. [Google Scholar] [CrossRef]
- Martin, W.; Russell, M.J. On the origin of biochemistry at an alkaline hydrothermal vent. Philos. Trans. R. Soc. B-Biol. Sci. 2007, 362, 1887–1925. [Google Scholar] [CrossRef] [PubMed]
- Colín-García, M.; Heredia, A.; Cordero, G.; Camprubí, A.; Negrón-Mendoza, A.; Ortega-Gutiérrez, F.; Beraldi, H.; Ramos-Bernal, S. Hydrothermal vents and prebiotic chemistry: A review. Boletín Soc. Geol. Mex. 2016, 68, 599–620. [Google Scholar] [CrossRef]
- Drobner, E.; Huber, H.; Wächtershäuser, G.; Rose, D.; Stetter, K.O. Pyrite formation linked with hydrogen evolution under anaerobic conditions. Nature 1990, 346, 742–744. [Google Scholar] [CrossRef]
- Martin, W.; Baross, J.; Kelley, D.; Russell, M.J. Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 2008, 6, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Sleep, N.H.; Bird, D.K.; Pope, E.C. Serpentinite and the dawn of life. Philos. Trans. R. Soc. B-Biol. Sci. 2011, 366, 2857–2869. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.C.; Sousa, F.L.; Mrnjavac, N.; Neukirchen, S.; Roettger, M.; Nelson-Sathi, S.; Martin, W.F. The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 2016, 1, 16116. [Google Scholar] [CrossRef]
- Reysenbach, A.L.; Shock, E. Merging genomes with geochemistry in hydrothermal ecosystems. Science 2002, 296, 1077–1082. [Google Scholar] [CrossRef]
- Kelley, D.S.; Karson, J.A.; Blackman, D.K.; Früh-Green, G.L.; Butterfield, D.A.; Lilley, M.D.; Olson, E.J.; Schrenk, M.O.; Roe, K.K.; Lebon, G.T.; et al. An off-axis hydrothermal vent field near the Mid-Atlantic Ridge at 30° N. Nature 2001, 412, 145–149. [Google Scholar] [CrossRef]
- Kelley, D.S.; Karson, J.A.; Früh-Green, G.L.; Yoerger, D.R.; Shank, T.M.; Butterfield, D.A.; Hayes, J.M.; Schrenk, M.O.; Olson, E.J.; Proskurowski, G.; et al. A serpentinite-hosted ecosystem: The lost city hydrothermal field. Science 2005, 307, 1428–1434. [Google Scholar] [CrossRef] [PubMed]
- Karson, J.A.; Früh-Green, G.L.; Kelley, D.S.; Williams, E.A.; Yoerger, D.R.; Jakuba, M. Detachment shear zone of the Atlantis Massif core complex, Mid-Atlantic Ridge, 30° N. Geochem. Geophys. Geosyst. 2006, 7, Q06016. [Google Scholar] [CrossRef]
- Preiner, M.; Igarashi, K.; Muchowska, K.B.; Yu, M.; Varma, S.J.; Kleinermanns, K.; Nobu, M.K.; Kamagata, Y.; Tüysüz, H.; Moran, J.; et al. A hydrogen-dependent geochemical analogue of primordial carbon and energy metabolism. Nat. Ecol. Evol. 2020, 4, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Russell, M.J.; Martin, W. The rocky roots of the acetyl-CoA pathway. Trends Biochem. Sci. 2004, 29, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Lane, N. Transformer; Profile Books: London, UK, 2023; p. 390. [Google Scholar]
- Muchowska, K.B.; Varma, S.J.; Moran, J. Nonenzymatic metabolic reactions and life’s origins. Chem. Rev. 2020, 120, 7708–7744. [Google Scholar] [CrossRef] [PubMed]
- Wächtershäuser, G. Groundworks for an evolutionary biochemistry: The iron-sulphur world. Prog. Biophys. Mol. Biol. 1992, 58, 85–201. [Google Scholar] [CrossRef] [PubMed]
- Russell, M.J. Green rust: The simple organizing ‘seed’ of all life? Life 2018, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Duval, S.; Branscomb, E.; Trolard, F.; Bourrié, G.; Grauby, O.; Heresanu, V.; Schoepp-Cothenet, B.; Zuchan, K.; Russell, M.J.; Nitschke, W. On the why’s and how’s of clay minerals’ importance in life’s emergence. Appl. Clay Sci. 2020, 195, 105737. [Google Scholar] [CrossRef]
- Xavier, J.C.; Gerhards, R.E.; Wimmer, J.L.E.; Brueckner, J.; Tria, F.D.K.; Martin, W.F. The metabolic network of the last bacterial common ancestor. Commun. Biol. 2021, 4, 413. [Google Scholar] [CrossRef]
- Martin, W.F. Older Than genes: The acetyl CoA pathway and origins. Front. Microbiol. 2020, 11, 817. [Google Scholar] [CrossRef]
- Xavier, J.C.; Hordijk, W.; Kauffman, S.; Steel, M.; Martin, W.F. Autocatalytic chemical networks at the origin of metabolism. Proc. R. Soc. B-Biol. Sci. 2020, 287, 20192377. [Google Scholar] [CrossRef] [PubMed]
- Braakman, R.; Smith, E. The emergence and early evolution of biological carbon fixation. PLoS Comput. Biol. 2012, 8, e1002455. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.A.; Kampjut, D.; Harrison, S.A.; Ralser, M. Sulfate radicals enable a non-enzymatic Krebs cycle precursor. Nat. Ecol. Evol. 2017, 1, 83. [Google Scholar] [CrossRef] [PubMed]
- Stubbs, R.T.; Yadav, M.; Krishnamurthy, R.; Springsteen, G. A plausible metal-free ancestral analogue of the Krebs cycle composed entirely of α-ketoacids. Nat. Chem. 2020, 12, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Noda-Garcia, L.; Liebermeister, W.; Tawfik, D.S. Metabolite-enzyme coevolution: From single enzymes to metabolic pathways and networks. Annu. Rev. Biochem. 2018, 87, 187–216. [Google Scholar] [CrossRef]
- Kim, K.M.; Caetano-Anollés, G. Emergence and evolution of modern molecular functions inferred from phylogenomic analysis of ontological data. Mol. Biol. Evol. 2010, 27, 1710–1733. [Google Scholar] [CrossRef] [PubMed]
- Fry, I. Emergence of Life on Earth: A Historical and Scientific Overview; Rutgers University Press: New Brunswick, NJ, USA, 2000. [Google Scholar]
- Copley, S.D.; Smith, E.; Morowitz, H.J. A mechanism for the association of amino acids with their codons and the origin of the genetic code. Proc. Natl. Acad. Sci. USA 2005, 102, 4442–4447. [Google Scholar] [CrossRef]
- Halpern, A.; Bartsch, L.R.; Ibrahim, K.; Harrison, S.A.; Ahn, M.; Christodoulou, J.; Lane, N. Biophysical interactions underpin the emergence of information in the genetic code. Life 2023, 13, 1129. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Opron, K.; Burton, Z.F. A tRNA- and anticodon-centric view of the evolution of aminoacyl-tRNA synthetases, tRNAomes, and the genetic code. Life 2019, 9, 37. [Google Scholar] [CrossRef]
- Pak, D.; Du, N.; Kim, Y.; Sun, Y.; Burton, Z.F. Rooted tRNAomes and evolution of the genetic code. Transcription 2018, 9, 137–151. [Google Scholar] [CrossRef]
- Lipmann, F. Attempts to map a process evolution of peptide biosynthesis. Science 1971, 173, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Wächtershäuser, G. The origin of life and its methodological challenge. J. Theor. Biol. 1997, 187, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Benner, S.A.; Ellington, A.D.; Tauer, A. Modern metabolism as a palimpsest of the RNA world. Proc. Natl. Acad. Sci. USA 1989, 86, 7054–7058. [Google Scholar] [CrossRef] [PubMed]
- Lyons, T.W.; Reinhard, C.T.; Planavsky, N.J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 2014, 506, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Meléndez-Hevia, E.; Montero-Gómez, N.; Montero, F. From prebiotic chemistry to cellular metabolism: The chemical evolution of metabolism before Darwinian natural selection. J. Theor. Biol. 2008, 252, 505–519. [Google Scholar] [CrossRef] [PubMed]
- Brazeau, M.D.; Ahlberg, P.E. Tetrapod-like middle ear architecture in a Devonian fish. Nature 2006, 439, 318–321. [Google Scholar] [CrossRef] [PubMed]
- Beeby, M.; Ferreira, J.L.; Tripp, P.; Albers, S.-V.; Mitchell, D.R. Propulsive nanomachines: The convergent evolution of archaella, flagella and cilia. FEMS Microbiol. Rev. 2020, 44, 253–304. [Google Scholar] [CrossRef]
- Heinen, W.; Lauwers, A.M. Organic sulfur compounds resulting from the interaction of iron sulfide, hydrogen sulfide and carbon dioxide in an anaerobic aqueous environment. Orig. Life Evol. Biosph. 1996, 26, 131–150. [Google Scholar] [CrossRef] [PubMed]
- Huber, C.; Wächtershäuser, G. Activated acetic acid by carbon fixation on (Fe,Ni)S under primordial conditions. Science 1997, 276, 245–247. [Google Scholar] [CrossRef]
- Brady, M.P.; Tostevin, R.; Tosca, N.J. Marine phosphate availability and the chemical origins of life on Earth. Nat. Commun. 2022, 13, 5162. [Google Scholar] [CrossRef]
- Schwartz, A.W. Phosphorus in prebiotic chemistry. Philos. Trans. R. Soc. B-Biol. Sci. 2006, 361, 1743–1749. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, B.; Muhling, J.R.; Suvorova, A.; Fischer, W.W. Apatite nanoparticles in 3.46-2.46 Ga iron formations: Evidence for phosphorus-rich hydrothermal plumes on early Earth. Geology 2021, 49, 647–651. [Google Scholar] [CrossRef]
- Kornberg, A.; Rao, N.N.; Ault-Riché, D. Inorganic polyphosphate: A molecule of many functions. Annu. Rev. Biochem. 1999, 68, 89–125. [Google Scholar] [CrossRef] [PubMed]
- Nam, I.; Lee, J.K.; Nam, H.G.; Zare, R.N. Abiotic production of sugar phosphates and uridine ribonucleoside in aqueous microdroplets. Proc. Natl. Acad. Sci. USA 2017, 114, 12396–12400. [Google Scholar] [CrossRef] [PubMed]
- Ralser, M. An appeal to magic? The discovery of a non-enzymatic metabolism and its role in the origins of life. Biochem. J. 2018, 475, 2577–2592. [Google Scholar] [CrossRef] [PubMed]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.-M.; Krüger, A.; Alam, M.T.; et al. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J.D. The origin of life—Out of the blue. Angew. Chem.-Int. Ed. 2016, 55, 104–121. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.; Pitsch, S.; Kittaka, A.; Wagner, E.; Wintner, C.E.; Eschenmoser, A. Chemistry of α-aminonitriles. Aldomerisation of glycolaldehyde phosphate to rac-hexose 2,4,6-triphosphates and (in presence of formaldehyde) rac-pentose 2,4-diphosphates: Rac-allose 2,4,6-triphosphate and rac-ribose2,4-diphosphate are the main reaction products. Helv. Chim. Acta 1990, 73, 1410–1468. [Google Scholar] [CrossRef]
- Yadav, M.; Kumar, R.; Krishnamurthy, R. Chemistry of abiotic nucleotide synthesis. Chem. Rev. 2020, 120, 4766–4805. [Google Scholar] [CrossRef]
- Srinivasan, V.; Morowitz, H.J. Analysis of the intermediary metabolism of a reductive chemoautotroph. Biol. Bull. 2009, 217, 222–232. [Google Scholar] [CrossRef]
- Dias, H.B.; Krause, G.C.; Squizani, E.D.; Lima, K.G.; Schuster, A.D.; Pedrazza, L.; Basso, B.d.S.; Martha, B.A.; de Mesquita, F.C.; Nunes, F.B.; et al. Fructose-1,6-bisphosphate reverts iron-induced phenotype of hepatic stellate cells by chelating ferrous ions. Biometals 2017, 30, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Swoboda, J.G.; Campbell, J.; Meredith, T.C.; Walker, S. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 2010, 11, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Damer, B.; Deamer, D. The hot spring hypothesis for an origin of life. Astrobiology 2020, 20, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Mollo, A.; Kahne, D.; Ruiz, N. The bacterial cell wall: From lipid II flipping to polymerization. Chem. Rev. 2022, 122, 8884–8910. [Google Scholar] [CrossRef] [PubMed]
- Dörr, M.; Kässbohrer, J.; Grunert, R.; Kreisel, G.; Brand, W.A.; Werner, R.A.; Geilmann, H.; Apfel, C.; Robl, C.; Weigand, W. A possible prebiotic formation of ammonia from dinitrogen on iron sulfide surfaces. Angew. Chem.-Int. Ed. 2003, 42, 1540–1543. [Google Scholar] [CrossRef] [PubMed]
- Huber, C.; Wächtershäuser, G. Primordial reductive amination revisited. Tetrahedron Lett. 2003, 44, 1695–1697. [Google Scholar] [CrossRef]
- Barge, L.M.; Flores, E.; Baum, M.M.; VanderVelde, D.G.; Russell, M.J. Redox and pH gradients drive amino acid synthesis in iron oxyhydroxide mineral systems. Proc. Natl. Acad. Sci. USA 2019, 116, 4828–4833. [Google Scholar] [CrossRef] [PubMed]
- Northrup, A.B.; MacMillan, D.W.C. Two-step synthesis of carbohydrates by selective aldol reactions. Science 2004, 305, 1752–1755. [Google Scholar] [CrossRef] [PubMed]
- List, B.; Pojarliev, P.; Biller, W.T.; Martin, H.J. The proline-catalyzed direct asymmetric three-component Mannich reaction: Scope, optimization, and application to the highly enantioselective synthesis of 1,2-amino alcohols. J. Am. Chem. Soc. 2002, 124, 827–833. [Google Scholar] [CrossRef]
- Messner, C.B.; Driscoll, P.C.; Piedrafita, G.; De Volder, M.F.L.; Ralser, M. Nonenzymatic gluconeogenesis-like formation of fructose 1,6-bisphosphate in ice. Proc. Natl. Acad. Sci. USA 2017, 114, 7403–7407. [Google Scholar] [CrossRef]
- Piedrafita, G.; Varma, S.J.; Castro, C.; Messner, C.; Szyrwiel, L.; Griffin, J.L.; Ralser, M. Cysteine and iron accelerate the formation of ribose-5-phosphate, providing insights into the evolutionary origins of the metabolic network structure. PLoS Biol. 2021, 19, e3001468. [Google Scholar] [CrossRef] [PubMed]
- Steer, A.M.; Bia, N.; Smith, D.K.; Clarke, P.A. Prebiotic synthesis of 2-deoxy-D-ribose from interstellar building blocks promoted by amino esters or amino nitriles. Chem. Commun. 2017, 53, 10362–10365. [Google Scholar] [CrossRef]
- Wächtershäuser, G. Biomolecules: The origin of their optical activity. Med. Hypotheses 1991, 36, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Breslow, R.; Cheng, Z.-L. L-amino acids catalyze the formation of an excess of D-glyceraldehyde, and thus of other D sugars, under credible prebiotic conditions. Proc. Natl. Acad. Sci. USA 2010, 107, 5723–5725. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.L.; Pizzarello, S. The peptide-catalyzed stereospecific synthesis of tetroses: A possible model for prebiotic molecular evolution. Proc. Natl. Acad. Sci. USA 2006, 103, 12713–12717. [Google Scholar] [CrossRef] [PubMed]
- Menor-Salván, C.; Burcar, B.T.; Bouza, M.; Fialho, D.M.; Fernández, F.M.; Hud, N.V. A shared prebiotic formation of neopterins and guanine nucleosides from pyrimidine bases. Chem.-A Eur. J. 2022, 28, e202200714. [Google Scholar] [CrossRef] [PubMed]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [PubMed]
- Kraus, W.L. PARPs and ADP-ribosylation: 50 years… and counting. Mol. Cell 2015, 58, 902–910. [Google Scholar] [CrossRef] [PubMed]
- White, H.B. Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 1976, 7, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T.; Tu, B.P.; Tane, Y. Eight kinetically stable but thermodynamically activated molecules that power cell metabolism. Chem. Rev. 2018, 118, 1460–1494. [Google Scholar] [CrossRef]
- Fournier, G.P.; Alm, E.J. Ancestral reconstruction of a pre-LUCA aminoacyl-tRNA synthetase ancestor supports the late addition of Trp to the genetic code. J. Mol. Evol. 2015, 80, 171–185. [Google Scholar] [CrossRef]
- Airapetian, V.S.; Glocer, A.; Gronoff, G.; Hébrard, E.; Danchi, W. Prebiotic chemistry and atmospheric warming of early Earth by an active young Sun. Nat. Geosci. 2016, 9, 452–455. [Google Scholar] [CrossRef]
- Aroskay, A.; Martin, E.; Bekki, S.; Le Pennec, J.-L.; Savarino, J.; Temel, A.; Manrique, N.; Aguilar, R.; Rivera, M.; Guillou, H.; et al. Geological evidence of extensive N-fixation by volcanic lightning during very large explosive eruptions. Proc. Natl. Acad. Sci. USA 2024, 121, e2309131121. [Google Scholar] [CrossRef]
- Oró, J. Synthesis of adenine from ammonium cyanide. Biochem. Biophys. Res. Commun. 1960, 2, 407–412. [Google Scholar] [CrossRef]
- Wu, L.-F.; Sutherland, J.D. Provisioning the origin and early evolution of life. Emerg. Top. Life Sci. 2019, 3, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.A.; Lane, N. Life as a guide to prebiotic nucleotide synthesis. Nat. Commun. 2018, 9, 5176. [Google Scholar] [CrossRef]
- Ferus, M.; Kubelík, P.; Knížek, A.; Pastorek, A.; Sutherland, J.; Civiš, S. High energy radical chemistry formation of HCN-rich atmospheres on early earth. Sci. Rep. 2017, 7, 6275. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.; Pulletikurti, S.; Yerabolu, J.R.; Krishnamurthy, R. Cyanide as a primordial reductant enables a protometabolic reductive glyoxylate pathway. Nat. Chem. 2022, 14, 170–178. [Google Scholar] [CrossRef]
- Petrov, A.S.; Gulen, B.; Norris, A.M.; Kovacs, N.A.; Bernier, C.R.; Lanier, K.A.; Fox, G.E.; Harvey, S.C.; Wartell, R.M.; Hud, N.V.; et al. History of the ribosome and the origin of translation. Proc. Natl. Acad. Sci. USA 2015, 112, 15396–15401. [Google Scholar] [CrossRef]
- Williamson, M.P. How Proteins Work; Garland Science: New York, NY, USA, 2011; p. 464. [Google Scholar]
- Söding, J.; Lupas, A.N. More than the sum of their parts: On the evolution of proteins from peptides. Bioessays 2003, 25, 837–846. [Google Scholar] [CrossRef]
- Dunkle, J.A.; Wang, L.; Feldman, M.B.; Pulk, A.; Chen, V.B.; Kapral, G.J.; Noeske, J.; Richardson, J.S.; Blanchard, S.C.; Cate, J.H.D. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 2011, 332, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Petrov, A.S.; Bernier, C.R.; Hsiao, C.; Norris, A.M.; Kovacs, N.A.; Waterbury, C.C.; Stepanov, V.G.; Harvey, S.C.; Fox, G.E.; Wartell, R.M.; et al. Evolution of the ribosome at atomic resolution. Proc. Natl. Acad. Sci. USA 2014, 111, 10251–10256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Will, C.L.; Bertram, K.; Dybkov, O.; Hartmuth, K.; Agafonov, D.E.; Hofele, R.; Urlaub, H.; Kastner, B.; Lührmann, R.; et al. Molecular architecture of the human 17S U2 snRNP. Nature 2020, 583, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.A.; Palmeira, R.N.; Halpern, A.; Lane, N. A biophysical basis for the emergence of the genetic code in protocells. Biochim. Biophys. Acta-Bioenerg. 2022, 1863, 148597. [Google Scholar] [CrossRef] [PubMed]
- Crick, F. Life Itself: Its Origin and Nature; Simon & Schuster: New York, NY, USA, 1981. [Google Scholar]



| Lipid vesicles able to catalyze (phospholipid) sugar polymerization? |
| Do phosphosugars form a barrier to diffusion of negatively charged pre-metabolites? |
| Polyphosphates as a route for producing sugar diphosphates. |
| Can HCN be produced in large amounts in Hadean conditions? |
| Are nitrogenous bases able to act as general nucleophilic catalysts? |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Williamson, M.P. Autocatalytic Selection as a Driver for the Origin of Life. Life 2024, 14, 590. https://doi.org/10.3390/life14050590
Williamson MP. Autocatalytic Selection as a Driver for the Origin of Life. Life. 2024; 14(5):590. https://doi.org/10.3390/life14050590
Chicago/Turabian StyleWilliamson, Mike P. 2024. "Autocatalytic Selection as a Driver for the Origin of Life" Life 14, no. 5: 590. https://doi.org/10.3390/life14050590






