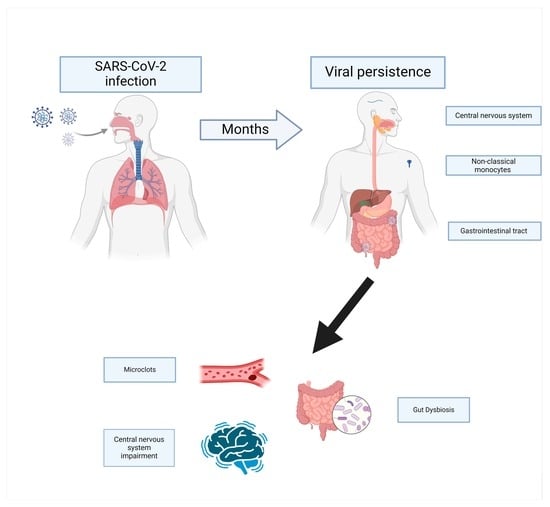
The Potential Role of Viral Persistence in the Post-Acute Sequelae of SARS-CoV-2 Infection (PASC)
Abstract
:1. Post-Acute Sequelae of SARS-CoV-2
2. Evidence of Persistence of SARS-CoV-2
2.1. Persistence of Viral Nucleic Acids and Antigens
2.2. Immune System-Based Evidence
2.2.1. Persistent Activation of the Immune System
2.2.2. Effects of Vaccination on PASC
2.2.3. Prolonged Evolution of Antibodies
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Salata, C.; Calistri, A.; Parolin, C.; Palù, G. Coronaviruses: A Paradigm of New Emerging Zoonotic Diseases. Pathog. Dis. 2019, 77, 6. [Google Scholar] [CrossRef]
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115. [Google Scholar] [CrossRef] [PubMed]
- Saloner, B.; Parish, K.; Julie Ward, M.A.; Grace DiLaura, R.; Sharon Dolovich, J. Persistent Symptoms in Patients After Acute COVID-19. JAMA 2020, 324, 603–605. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Haupert, S.R.; Zimmermann, L.; Shi, X.; Fritsche, L.G.; Mukherjee, B. Global Prevalence of Post-Coronavirus Disease 2019 (COVID-19) Condition or Long COVID: A Meta-Analysis and Systematic Review. J. Infect. Dis. 2022, 226, 1593–1607. [Google Scholar] [CrossRef] [PubMed]
- Available online: Https://Www.Who.Int/Publications/i/Item/WHO-2019-NCoV-Post_COVID-19_condition-Clinical_case_definition-2021.1 (accessed on 5 May 2024).
- Thaweethai, T.; Jolley, S.E.; Karlson, E.W.; Levitan, E.B.; Levy, B.; McComsey, G.A.; McCorkell, L.; Nadkarni, G.N.; Parthasarathy, S.; Singh, U.; et al. Development of a Definition of Postacute Sequelae of SARS-CoV-2 Infection. JAMA 2023, 329, 1934–1946. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Huang, L.; Wang, Y.; Li, X.; Ren, L.; Gu, X.; Kang, L.; Guo, L.; Liu, M.; Zhou, X.; et al. 6-Month Consequences of COVID-19 in Patients Discharged from Hospital: A Cohort Study. Lancet 2021, 397, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Blomberg, B.; Mohn, K.G.I.; Brokstad, K.A.; Zhou, F.; Linchausen, D.W.; Hansen, B.A.; Lartey, S.; Onyango, T.B.; Kuwelker, K.; Sævik, M.; et al. Long COVID in a Prospective Cohort of Home-Isolated Patients. Nat. Med. 2021, 27, 1607–1613. [Google Scholar] [CrossRef] [PubMed]
- Yong, S.J. Long COVID or Post-COVID-19 Syndrome: Putative Pathophysiology, Risk Factors, and Treatments. Infect. Dis. 2021, 53, 737–754. [Google Scholar] [CrossRef]
- Spudich, S.; Nath, A. Nervous System Consequences of COVID-19. Science 2022, 375, 267–269. [Google Scholar] [CrossRef]
- Perumal, R.; Shunmugam, L.; Naidoo, K.; Abdool Karim, S.S.; Wilkins, D.; Garzino-Demo, A.; Brechot, C.; Parthasarathy, S.; Vahlne, A.; Nikolich, J. Long COVID: A Review and Proposed Visualization of the Complexity of Long COVID. Front. Immunol. 2023, 14, 1117464. [Google Scholar] [CrossRef] [PubMed]
- Perumal, R.; Shunmugam, L.; Naidoo, K.; Wilkins, D.; Garzino-Demo, A.; Brechot, C.; Vahlne, A.; Nikolich, J. Biological Mechanisms Underpinning the Development of Long COVID. iScience 2023, 26, 106935. [Google Scholar] [CrossRef] [PubMed]
- Tsampasian, V.; Elghazaly, H.; Chattopadhyay, R.; Debski, M.; Naing, T.K.P.; Garg, P.; Clark, A.; Ntatsaki, E.; Vassiliou, V.S. Risk Factors Associated With Post-COVID-19 Condition A Systematic Review and Meta-Analysis. JAMA Intern. Med. 2023, 183, 566–580. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B.; Laubscher, G.J.; Pretorius, E. A Central Role for Amyloid Fibrin Microclots in Long COVID/PASC: Origins and Therapeutic Implications. Biochem. J. 2022, 479, 537–559. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Wu, X.; Xiang, M.; Liu, L.; Novakovic, V.A.; Shi, J. Pathophysiological Mechanisms of Thrombosis in Acute and Long COVID-19. Front. Immunol. 2022, 13, 992384. [Google Scholar] [CrossRef]
- Greene, C.; Connolly, R.; Brennan, D.; Laffan, A.; O’Keeffe, E.; Zaporojan, L.; O’Callaghan, J.; Thomson, B.; Connolly, E.; Argue, R.; et al. Blood–Brain Barrier Disruption and Sustained Systemic Inflammation in Individuals with Long COVID-Associated Cognitive Impairment. Nat. Neurosci. 2024, 27, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Castañeda, A.; Lu, P.; Geraghty, A.C.; Song, E.; Lee, M.-H.; Wood, J.; O’Dea, M.R.; Dutton, S.; Shamardani, K.; Nwangwu, K.; et al. Mild Respiratory COVID Can Cause Multi-Lineage Neural Cell and Myelin Dysregulation. Cell 2022, 185, 2452–2468. [Google Scholar] [CrossRef] [PubMed]
- Dotan, A.; Muller, S.; Kanduc, D.; David, P.; Halpert, G.; Shoenfeld, Y. The SARS-CoV-2 as an Instrumental Trigger of Autoimmunity. Autoimmun. Rev. 2021, 20, 102792. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, A.C.; Humbert, M.; Buggert, M. The Known Unknowns of T Cell Immunity to COVID-19. Sci. Immunol. 2020, 5, 8063. [Google Scholar] [CrossRef]
- Chang, S.E.; Feng, A.; Meng, W.; Apostolidis, S.A.; Mack, E.; Artandi, M.; Barman, L.; Bennett, K.; Chakraborty, S.; Chang, I.; et al. New-Onset IgG Autoantibodies in Hospitalized Patients with COVID-19. Nat. Commun. 2021, 12, 5417. [Google Scholar] [CrossRef]
- Apostolou, E.; Rizwan, M.; Moustardas, P.; Sjögren, P.; Bertilson, B.C.; Bragée, B.; Polo, O.; Rosén, A. Saliva Antibody-Fingerprint of Reactivated Latent Viruses after Mild/Asymptomatic COVID-19 Is Unique in Patients with Myalgic-Encephalomyelitis/Chronic Fatigue Syndrome. Front. Immunol. 2022, 13, 949787. [Google Scholar] [CrossRef]
- Gold, J.E.; Okyay, R.A.; Licht, W.E.; Hurley, D.J. Investigation of Long Covid Prevalence and Its Relationship to Epstein-Barr Virus Reactivation. Pathogens 2021, 10, 763. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Leon, S.; Wegman-Ostrosky, T.; Perelman, C.; Sepulveda, R.; Rebolledo, P.A.; Cuapio, A.; Villapol, S. More than 50 Long-Term Effects of COVID-19: A Systematic Review and Meta-Analysis. Sci. Rep. 2021, 11, 16144. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, M.; Pujol, J.C.; Spector, T.D.; Ourselin, S.; Steves, C.J. Risk of Long COVID Associated with Delta versus Omicron Variants of SARS-CoV-2. Lancet 2022, 399, 2263–2264. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.A.; Morden, E.; Rousseau, P.; Arendse, J.; Bam, J.L.; Boloko, L.; Cloete, K.; Cohen, C.; Chetty, N.; Dane, P.; et al. Outcomes of Laboratory-Confirmed SARS-CoV-2 Infection during Resurgence Driven by Omicron Lineages BA.4 and BA.5 Compared with Previous Waves in the Western Cape Province, South Africa. Int. J. Infect. Dis. 2023, 127, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Cegolon, L.; Mauro, M.; Sansone, D.; Tassinari, A.; Gobba, F.M.; Modenese, A.; Casolari, L.; Liviero, F.; Pavanello, S.; Scapellato, M.L.; et al. A Multi-Center Study Investigating Long COVID-19 in Healthcare Workers from North-Eastern Italy: Prevalence, Risk Factors and the Impact of Pre-Existing Humoral Immunity—ORCHESTRA Project. Vaccines 2023, 11, 1769. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, P.; Russo, A.; Pisaturo, M.; Maggi, P.; Allegorico, E.; Gentile, I.; Sangiovanni, V.; Rossomando, A.; Pacilio, R.; Calabria, G.; et al. Clinical and Epidemiological Factors Causing Longer SARS-CoV 2 Viral Shedding: The Results from the CoviCamp Cohort. Infection 2023, 52, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Seo, Y.B.; Seo, J.W.; Lee, J.; Nham, E.; Seong, H.; Yoon, J.G.; Noh, J.Y.; Cheong, H.J.; Kim, W.J.; et al. Effectiveness of Antiviral Therapy on Long COVID: A Systematic Review and Meta-Analysis. J. Clin. Med. 2023, 12, 7375. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Choi, T.; Al-Aly, Z. Molnupiravir and Risk of Post-Acute Sequelae of COVID-19: Cohort Study. BMJ 2023, 381, e074572. [Google Scholar] [CrossRef]
- Xie, Y.; Choi, T.; Al-Aly, Z. Association of Treatment With Nirmatrelvir and the Risk of Post-COVID-19 Condition. JAMA Intern. Med. 2023, 183, 554–564. [Google Scholar] [CrossRef]
- Lasagna, A.; Albi, G.; Figini, S.; Basile, S.; Sacchi, P.; Bruno, R.; Pedrazzoli, P. Long-COVID in Patients with Cancer Previously Treated with Early Anti-SARS-CoV-2 Therapies in an Out-of-Hospital Setting: A Single-Center Experience. Cancers 2023, 15, 1269. [Google Scholar] [CrossRef] [PubMed]
- Zapor, M. Persistent Detection and Infectious Potential of SARS-CoV-2 Virus in Clinical Specimens from COVID-19 Patients. Viruses 2020, 12, 1384. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.L.H.; Raval, M.; Schnall, J.A.; Kwong, J.C.; Holmes, N.E. Duration of Respiratory and Gastrointestinal Viral Shedding in Children with SARS-CoV-2: A Systematic Review and Synthesis of Data. Pediatr. Infect. Dis. J. 2020, 39, E249–E256. [Google Scholar] [CrossRef] [PubMed]
- Kipkorir, V.; Cheruiyot, I.; Ngure, B.; Misiani, M.; Munguti, J. Prolonged SARS-CoV-2 RNA Detection in Anal/Rectal Swabs and Stool Specimens in COVID-19 Patients after Negative Conversion in Nasopharyngeal RT-PCR Test. J. Med. Virol. 2020, 92, 2328–2331. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Xie, Z.; Li, Y.; Li, L.; Wen, C.; Cao, Y.; Chen, X.; Ou, X.; Hu, F.; Li, F.; et al. Association between Detectable SARS-COV-2 RNA in Anal Swabs and Disease Severity in Patients with Coronavirus Disease 2019. J. Med. Virol. 2021, 93, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, A.; Zlitni, S.; Brooks, E.F.; Vance, S.E.; Dahlen, A.; Hedlin, H.; Park, R.M.; Han, A.; Schmidtke, D.T.; Verma, R.; et al. Gastrointestinal Symptoms and Fecal Shedding of SARS-CoV-2 RNA Suggest Prolonged Gastrointestinal Infection. Med 2022, 3, 371–387.e9. [Google Scholar] [CrossRef]
- Zollner, A.; Koch, R.; Jukic, A.; Pfister, A.; Meyer, M.; Rössler, A.; Kimpel, J.; Adolph, T.E.; Tilg, H. Postacute COVID-19 Is Characterized by Gut Viral Antigen Persistence in Inflammatory Bowel Diseases. Gastroenterology 2022, 163, 495–506. [Google Scholar] [CrossRef]
- Stein, S.R.; Ramelli, S.C.; Grazioli, A.; Chung, J.Y.; Singh, M.; Yinda, C.K.; Winkler, C.W.; Sun, J.; Dickey, J.M.; Ylaya, K.; et al. SARS-CoV-2 Infection and Persistence in the Human Body and Brain at Autopsy. Nature 2022, 612, 758–763. [Google Scholar] [CrossRef]
- Yao, Q.; Doyle, M.E.; Liu, Q.-R.; Appleton, A.; O’Connell, J.F.; Weng, N.; Egan, J.M. Long-Term Dysfunction of Taste Papillae in SARS-CoV-2. NEJM Evid. 2023, 22, EVIDoa2300046. [Google Scholar] [CrossRef]
- Rong, Z.; Mai, H.; Kapoor, S.; Puelles, V.G.; Czogalla, J.; Schädler, J.; Vering, J.; Delbridge, C.; Steinke, H.; Frenzel, H.; et al. SARS-CoV-2 Spike Protein Accumulation in the Skull-Meninges-Brain Axis: Potential Implications for Long-Term Neurological Complications in Post-COVID-19. BioRxiv 2023, bioRxiv 2023.04.04.535604. [Google Scholar] [CrossRef]
- Huot, N.; Planchais, C.; Rosenbaum, P.; Contreras, V.; Jacquelin, B.; Petitdemange, C.; Lazzerini, M.; Beaumont, E.; Orta-Resendiz, A.; Rey, F.A.; et al. SARS-CoV-2 Viral Persistence in Lung Alveolar Macrophages Is Controlled by IFN-γ and NK Cells. Nat. Immunol. 2023, 24, 2068–2079. [Google Scholar] [CrossRef] [PubMed]
- Patterson, B.K.; Francisco, E.B.; Yogendra, R.; Long, E.; Pise, A.; Rodrigues, H.; Hall, E.; Herrera, M.; Parikh, P.; Guevara-Coto, J.; et al. Persistence of SARS CoV-2 S1 Protein in CD16+ Monocytes in Post-Acute Sequelae of COVID-19 (PASC) up to 15 Months Post-Infection. Front. Immunol. 2022, 12, 746021. [Google Scholar] [CrossRef] [PubMed]
- Junqueira, C.; Crespo, Â.; Ranjbar, S.; de Lacerda, L.B.; Lewandrowski, M.; Ingber, J.; Parry, B.; Ravid, S.; Clark, S.; Schrimpf, M.R.; et al. FcγR-Mediated SARS-CoV-2 Infection of Monocytes Activates Inflammation. Nature 2022, 606, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Lupi, L.; Bordin, A.; Sales, G.; Colaianni, D.; Vitiello, A.; Biscontin, A.; Reale, A.; Garzino-Demo, A.; Antonini, A.; Ottaviano, G.; et al. Persistent and Transient Olfactory Deficits in COVID-19 Are Associated to Inflammation and Zinc Homeostasis. Front. Immunol. 2023, 14, 1148595. [Google Scholar] [CrossRef] [PubMed]
- Frere, J.J.; Serafini, R.A.; Pryce, K.D.; Zazhytska, M.; Oishi, K.; Golynker, I.; Panis, M.; Zimering, J.; Horiuchi, S.; Hoagland, D.A.; et al. SARS-CoV-2 Infection in Hamsters and Humans Results in Lasting and Unique Systemic Perturbations Post Recovery. Sci. Transl. Med. 2022, 14, eabq3059. [Google Scholar] [CrossRef]
- Phetsouphanh, C.; Darley, D.R.; Wilson, D.B.; Howe, A.; Munier, C.M.L.; Patel, S.K.; Juno, J.A.; Burrell, L.M.; Kent, S.J.; Dore, G.J.; et al. Immunological Dysfunction Persists for 8 Months Following Initial Mild-to-Moderate SARS-CoV-2 Infection. Nat. Immunol. 2022, 23, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Paniskaki, K.; Goretzki, S.; Anft, M.; Konik, M.J.; Meister, T.L.; Pfaender, S.; Lechtenberg, K.; Vogl, M.; Dogan, B.; Dolff, S.; et al. Increased SARS-CoV-2 Reactive Low Avidity T Cells Producing Inflammatory Cytokines in Pediatric Post-Acute COVID-19 Sequelae (PASC). Pediatr. Allergy Immunol. 2023, 34, e14060. [Google Scholar] [CrossRef] [PubMed]
- Littlefield, K.M.; Watson, R.O.; Schneider, J.M.; Neff, C.P.; Yamada, E.; Zhang, M.; Campbell, T.B.; Falta, M.T.; Jolley, S.E.; Fontenot, A.P.; et al. SARS-CoV-2-Specific T Cells Associate with Inflammation and Reduced Lung Function in Pulmonary Post-Acute Sequalae of SARS-CoV-2. PLoS Pathog. 2022, 18, e1010359. [Google Scholar] [CrossRef] [PubMed]
- Visvabharathy, L.; Hanson, B.A.; Orban, Z.S.; Lim, P.H.; Palacio, N.M.; Jimenez, M.; Clark, J.R.; Graham, E.L.; Liotta, E.M.; Tachas, G.; et al. Neuro-PASC Is Characterized by Enhanced CD4+ and Diminished CD8+ T Cell Responses to SARS-CoV-2 Nucleocapsid Protein. Front. Immunol. 2023, 14, 1155770. [Google Scholar] [CrossRef]
- Yin, K.; Peluso, M.J.; Luo, X.; Thomas, R.; Shin, M.-G.; Neidleman, J.; Andrew, A.; Young, K.C.; Ma, T.; Hoh, R.; et al. Long COVID Manifests with T Cell Dysregulation, Inflammation and an Uncoordinated Adaptive Immune Response to SARS-CoV-2. Nat. Immunol. 2024, 25, 218–225. [Google Scholar] [CrossRef]
- Santa Cruz, A.; Mendes-Frias, A.; Azarias-da-Silva, M.; André, S.; Oliveira, A.I.; Pires, O.; Mendes, M.; Oliveira, B.; Braga, M.; Lopes, J.R.; et al. Post-Acute Sequelae of COVID-19 Is Characterized by Diminished Peripheral CD8+β7 Integrin+ T Cells and Anti-SARS-CoV-2 IgA Response. Nat. Commun. 2023, 14, 1772. [Google Scholar] [CrossRef] [PubMed]
- Cervia-Hasler, C.; Brüningk, S.C.; Hoch, T.; Fan, B.; Muzio, G.; Thompson, R.C.; Ceglarek, L.; Meledin, R.; Westermann, P.; Emmenegger, M.; et al. Persistent Complement Dysregulation with Signs of Thromboinflammation in Active Long Covid. Science 2024, 383, eadg7942. [Google Scholar] [CrossRef] [PubMed]
- Ledford, H. Do Vaccines Protect against Long COVID? What the Data Say. Nature 2021, 599, 546–548. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, M.; Penfold, R.S.; Merino, J.; Sudre, C.H.; Molteni, E.; Berry, S.; Canas, L.S.; Graham, M.S.; Klaser, K.; Modat, M.; et al. Risk Factors and Disease Profile of Post-Vaccination SARS-CoV-2 Infection in UK Users of the COVID Symptom Study App: A Prospective, Community-Based, Nested, Case-Control Study. Lancet Infect. Dis. 2022, 22, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Lam, I.C.H.; Zhang, R.; Man, K.K.C.; Wong, C.K.H.; Chui, C.S.L.; Lai, F.T.T.; Li, X.; Chan, E.W.Y.; Lau, C.S.; Wong, I.C.K.; et al. Persistence in Risk and Effect of COVID-19 Vaccination on Long-Term Health Consequences after SARS-CoV-2 Infection. Nat. Commun. 2024, 15, 1716. [Google Scholar] [CrossRef] [PubMed]
- Català, M.; Mercadé-Besora, N.; Kolde, R.; Trinh, N.T.H.; Roel, E.; Burn, E.; Rathod-Mistry, T.; Kostka, K.; Man, W.Y.; Delmestri, A.; et al. The Effectiveness of COVID-19 Vaccines to Prevent Long COVID Symptoms: Staggered Cohort Study of Data from the UK, Spain, and Estonia. Lancet Respir. Med. 2024, 12, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Lundberg-Morris, L.; Leach, S.; Xu, Y.; Martikainen, J.; Santosa, A.; Gisslén, M.; Li, H.; Nyberg, F.; Bygdell, M. COVID-19 Vaccine Effectiveness against Post-COVID-19 Condition among 589 722 Individuals in Sweden: Population Based Cohort Study. BMJ 2023, 383, e076990. [Google Scholar] [CrossRef] [PubMed]
- Coronavirus (COVID-19) Vaccination and Self-Reported Long COVID in the UK—Office for National Statistics. Available online: https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/articles/coronaviruscovid19vaccinationandselfreportedlongcovidintheuk/25october2021 (accessed on 27 December 2022).
- Strain, W.D.; Sherwood, O.; Banerjee, A.; van der Togt, V.; Hishmeh, L.; Rossman, J. The Impact of COVID Vaccination on Symptoms of Long COVID: An International Survey of People with Lived Experience of Long COVID. Vaccines 2022, 10, 652. [Google Scholar] [CrossRef] [PubMed]
- Ayoubkhani, D.; Bermingham, C.; Pouwels, K.B.; Glickman, M.; Nafilyan, V.; Zaccardi, F.; Khunti, K.; Alwan, N.A.; Walker, A.S. Trajectory of Long Covid Symptoms after COVID-19 Vaccination: Community Based Cohort Study. BMJ 2022, 377, e069676. [Google Scholar] [CrossRef]
- Marra, A.R.; Kobayashi, T.; Callado, G.Y.; Pardo, I.; Gutfreund, M.C.; Hsieh, M.K.; Lin, V.; Alsuhaibani, M.; Hasegawa, S.; Tholany, J.; et al. The Effectiveness of COVID-19 Vaccine in the Prevention of Post-COVID Conditions: A Systematic Literature Review and Meta-Analysis of the Latest Research. Antimicrob. Steward. Healthc. Epidemiol. 2023, 3, e168. [Google Scholar] [CrossRef]
- Hwang, J.K.; Alt, F.W.; Yeap, L.-S. Related Mechanisms of Antibody Somatic Hypermutation and Class Switch Recombination. Microbiol. Spectr. 2015, 3, 325–348. [Google Scholar] [CrossRef]
- Wu, J.; Chen, Z.; Gao, Y.; Wang, Z.; Wang, J.; Chiang, B.-Y.; Zhou, Y.; Han, Y.; Zhan, W.; Xie, M.; et al. Fortuitous Somatic Mutations during Antibody Evolution Endow Broad Neutralization against SARS-CoV-2 Omicron Variants. Cell Rep. 2023, 42, 112503. [Google Scholar] [CrossRef]
- Gaebler, C.; Wang, Z.; Lorenzi, J.C.C.; Muecksch, F.; Finkin, S.; Tokuyama, M.; Cho, A.; Jankovic, M.; Schaefer-Babajew, D.; Oliveira, T.Y.; et al. Evolution of Antibody Immunity to SARS-CoV-2. Nature 2021, 591, 639–644. [Google Scholar] [CrossRef]
- Files, J.K.; Sarkar, S.; Fram, T.R.; Boppana, S.; Sterrett, S.; Qin, K.; Bansal, A.; Long, D.M.; Sabbaj, S.; Kobie, J.J.; et al. Duration of Post–COVID-19 Symptoms Is Associated with Sustained SARS-CoV-2–Specific Immune Responses. JCI Insight 2021, 6, e151544. [Google Scholar] [CrossRef]
- Xu, Q.; Milanez-Almeida, P.; Martins, A.J.; Radtke, A.J.; Hoehn, K.B.; Oguz, C.; Chen, J.; Liu, C.; Tang, J.; Grubbs, G.; et al. Adaptive Immune Responses to SARS-CoV-2 Persist in the Pharyngeal Lymphoid Tissue of Children. Nat. Immunol. 2022, 24, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Bestagno, M.; Burrone, O.; Petrini, M. CD57+ T Lymphocytes and Functional Immune Deficiency. J. Leukoc. Biol. 2010, 87, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Sokal, A.; Chappert, P.; Barba-Spaeth, G.; Roeser, A.; Fourati, S.; Azzaoui, I.; Vandenberghe, A.; Fernandez, I.; Meola, A.; Bouvier-Alias, M.; et al. Maturation and Persistence of the Anti-SARS-CoV-2 Memory B Cell Response. Cell 2021, 184, 1201–1213.e14. [Google Scholar] [CrossRef]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major Findings, Mechanisms and Recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef]
- Chiang, C.H.; Shih, J.F.; Su, W.J.; Perng, R.P. Eight-Month Prospective Study of 14 Patients with Hospital-Acquired Severe Acute Respiratory Syndrome. Mayo Clin. Proc. 2004, 79, 1372–1379. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.S.; Zheng, J.P.; Mok, Y.W.; Li, Y.M.; Liu, Y.N.; Chu, C.M.; Ip, M.S. SARS: Prognosis, Outcome and Sequelae. Respirology 2003, 8, S36–S40. [Google Scholar] [CrossRef]
- Ahmed, H.; Patel, K.; Greenwood, D.C.; Halpin, S.; Lewthwaite, P.; Salawu, A.; Eyre, L.; Breen, A.; O’Connor, R.; Jones, A.; et al. Long-Term Clinical Outcomes in Survivors of Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) Coronavirus Outbreaks after Hospitalisation or ICU Admission: A Systematic Review and Meta-Analysis. J. Rehabil. Med. 2020, 52, 1–11. [Google Scholar] [CrossRef]
- Griffin, D.E. Why Does Viral RNA Sometimes Persist after Recovery from Acute Infections? PLoS Biol. 2022, 20, e3001687. [Google Scholar] [CrossRef]
- Burki, T.K. Post-Ebola Syndrome. Lancet Infect. Dis. 2016, 16, 780–781. [Google Scholar] [CrossRef]
- Scott, J.T.; Sesay, F.R.; Massaquoi, T.A.; Idriss, B.R.; Sahr, F.; Semple, M.G. Post-Ebola Syndrome, Sierra Leone. Emerg. Infect. Dis. 2016, 22, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Bond, N.G.; Grant, D.S.; Himmelfarb, S.T.; Engel, E.J.; Al-Hasa, F.; Gbakie, M.; Kamara, F.; Kanneh, L.; Mustapha, I.; Okoli, A.; et al. Post-Ebola Syndrome Presents With Multiple Overlapping Symptom Clusters: Evidence From an Ongoing Cohort Study in Eastern Sierra Leone. Clin. Infect. Dis. 2021, 73, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Ozonoff, A.; Jayavelu, N.D.; Liu, S.; Melamed, E.; Milliren, C.E.; Qi, J.; Geng, L.N.; McComsey, G.A.; Cairns, C.B.; Baden, L.R.; et al. Features of Acute COVID-19 Associated with Post-Acute Sequelae of SARS-CoV-2 Phenotypes: Results from the IMPACC Study. Nat. Commun. 2024, 15, 216. [Google Scholar] [CrossRef]
- Giannos, P.; Prokopidis, K. Gut Dysbiosis and Long COVID-19: Feeling Gutted. J. Med. Virol. 2022, 94, 2917–2918. [Google Scholar] [CrossRef]
- Haran, J.P.; Bradley, E.; Zeamer, A.L.; Cincotta, L.; Salive, M.C.; Dutta, P.; Mutaawe, S.; Anya, O.; Meza-Segura, M.; Moormann, A.M.; et al. Inflammation-Type Dysbiosis of the Oral Microbiome Associates with the Duration of COVID-19 Symptoms and Long COVID. JCI Insight 2021, 6, e152346. [Google Scholar] [CrossRef]
- Bicknell, B.; Liebert, A.; Borody, T.; Herkes, G.; McLachlan, C.; Kiat, H. Neurodegenerative and Neurodevelopmental Diseases and the Gut-Brain Axis: The Potential of Therapeutic Targeting of the Microbiome. Int. J. Mol. Sci. 2023, 24, 9577. [Google Scholar] [CrossRef]
- Wang, T.; Pan, C.; Xie, C.; Chen, L.; Song, Z.; Liao, H.; Xin, C. Microbiota Metabolites and Immune Regulation Affect Ischemic Stroke Occurrence, Development, and Prognosis. Mol. Neurobiol. 2023, 60, 6176–6187. [Google Scholar] [CrossRef]
- Naufel, M.F.; de Martin Truzzi, G.; Ferreira, C.M.; Coelho, F.M.S. The Brain-Gut-Microbiota Axis in the Treatment of Neurologic and Psychiatric Disorders. Arq. Neuropsiquiatr. 2023, 81, 670–684. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.C.; Devason, A.S.; Umana, I.C.; Cox, T.O.; Dohnalová, L.; Litichevskiy, L.; Perla, J.; Lundgren, P.; Etwebi, Z.; Izzo, L.T.; et al. Serotonin Reduction in Post-Acute Sequelae of Viral Infection. Cell 2023, 186, 4851–4867. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Song, Y.; Xu, Y.; Xu, H. Manipulating Microbiota in Inflammatory Bowel Disease Treatment: Clinical and Natural Product Interventions Explored. Int. J. Mol. Sci. 2023, 24, 11004. [Google Scholar] [CrossRef] [PubMed]
- Grimstad, T.; Norheim, K.B.; Isaksen, K.; Leitao, K.; Hetta, A.K.; Carlsen, A.; Karlsen, L.N.; Skoie, I.M.; Gøransson, L.; Harboe, E.; et al. Fatigue in Newly Diagnosed Inflammatory Bowel Disease. J. Crohns. Colitis 2015, 9, 725–730. [Google Scholar] [CrossRef]
- Liu, Q.; Mak, J.W.Y.; Su, Q.; Yeoh, Y.K.; Lui, G.C.Y.; Ng, S.S.S.; Zhang, F.; Li, A.Y.L.; Lu, W.; Hui, D.S.C.; et al. Gut Microbiota Dynamics in a Prospective Cohort of Patients with Post-Acute COVID-19 Syndrome. Gut 2022, 71, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, V.L.; Littlefield, K.M.; Watson, R.; Palmer, B.E.; Lozupone, C. Inflammation-Associated Gut Microbiome in Postacute Sequelae of SARS-CoV-2 Points towards New Therapeutic Targets. Gut 2023, 73, 376–378. [Google Scholar] [CrossRef] [PubMed]
- Kusakabe, T.; Lin, W.Y.; Cheong, J.G.; Singh, G.; Ravishankar, A.; Yeung, S.T.; Mesko, M.; DeCelie, M.B.; Carriche, G.; Zhao, Z.; et al. Fungal Microbiota Sustains Lasting Immune Activation of Neutrophils and Their Progenitors in Severe COVID-19. Nat. Immunol. 2023, 24, 1879–1889. [Google Scholar] [CrossRef]
- Wolf, A.A.; Yáñez, A.; Barman, P.K.; Goodridge, H.S. The Ontogeny of Monocyte Subsets. Front. Immunol. 2019, 10, 1642. [Google Scholar] [CrossRef]
- Klein, J.; Wood, J.; Jaycox, J.R.; Dhodapkar, R.M.; Lu, P.; Gehlhausen, J.R.; Tabachnikova, A.; Greene, K.; Tabacof, L.; Malik, A.A.; et al. Distinguishing Features of Long COVID Identified through Immune Profiling. Nature 2023, 623, 139–148. [Google Scholar] [CrossRef]
- Cho, S.M.; Premraj, L.; Battaglini, D.; Fanning, J.P.; Suen, J.; Bassi, G.L.; Fraser, J.; Robba, C.; Griffee, M.; Solomon, T.; et al. Sex Differences in Post-Acute Neurological Sequelae of SARS-CoV-2 and Symptom Resolution in Adults after Coronavirus Disease 2019 Hospitalization: An International Multi-Centre Prospective Observational Study. Brain Commun. 2024, 6, fcae036. [Google Scholar] [CrossRef]
- Takahashi, T.; Ellingson, M.K.; Wong, P.; Israelow, B.; Lucas, C.; Klein, J.; Silva, J.; Mao, T.; Oh, J.E.; Tokuyama, M.; et al. Sex Differences in Immune Responses That Underlie COVID-19 Disease Outcomes. Nature 2020, 588, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Roessler, M.; Tesch, F.; Batram, M.; Jacob, J.; Loser, F.; Weidinger, O.; Wende, D.; Vivirito, A.; Toepfner, N.; Ehm, F.; et al. Post-COVID-19 -Associated Morbidity in Children, Adolescents, and Adults: A Matched Cohort Study Including More than 157,000 Individuals with COVID-19 in Germany. PLoS Med. 2022, 19, e1004122. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Li, X.; Nie, J.; Tang, K.; Bhutta, Z.A. A Systematic Review of Persistent Clinical Features After SARS-CoV-2 in the Pediatric Population. Pediatrics 2023, 152, 2022060351. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lupi, L.; Vitiello, A.; Parolin, C.; Calistri, A.; Garzino-Demo, A. The Potential Role of Viral Persistence in the Post-Acute Sequelae of SARS-CoV-2 Infection (PASC). Pathogens 2024, 13, 388. https://doi.org/10.3390/pathogens13050388
Lupi L, Vitiello A, Parolin C, Calistri A, Garzino-Demo A. The Potential Role of Viral Persistence in the Post-Acute Sequelae of SARS-CoV-2 Infection (PASC). Pathogens. 2024; 13(5):388. https://doi.org/10.3390/pathogens13050388
Chicago/Turabian StyleLupi, Lorenzo, Adriana Vitiello, Cristina Parolin, Arianna Calistri, and Alfredo Garzino-Demo. 2024. "The Potential Role of Viral Persistence in the Post-Acute Sequelae of SARS-CoV-2 Infection (PASC)" Pathogens 13, no. 5: 388. https://doi.org/10.3390/pathogens13050388