Genotypic and Phenotypic Characteristics of Lactic Acid Bacteria Associated with Forage Plants in the Native Grassland of Western Inner Mongolia and Their Application for Alfalfa Silage Fermentation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and LAB Isolation
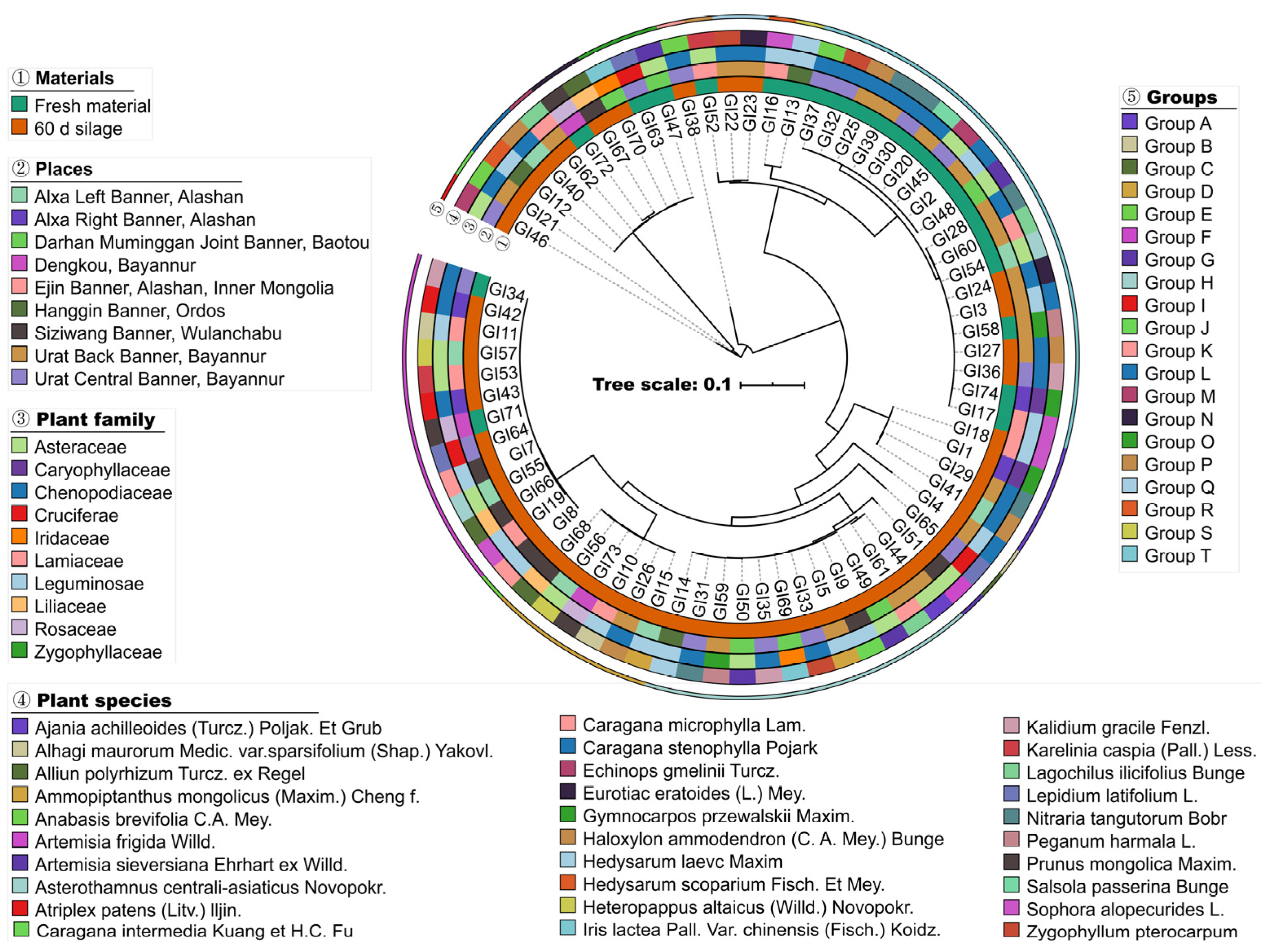
2.2. Phylogenetic Relationships among Isolates
2.3. Morphological, Physiological, and Biochemical Tests of LAB
2.4. Species Identification by 16S rDNA Sequencing, pheS and rpoA Genes
2.5. Silage Preparation and Microbial and Chemical Analysis
2.6. Statistical Analysis
3. Results
3.1. Isolation of LAB
3.2. The Morphological, Physiological, and Biochemical Properties of LAB Strains
3.3. Phylogenetic Analysis of 16S rDNA Sequence
3.4. Phylogenetic Analysis of pheS and rpoA Genes
3.5. Fermentation Characteristics of Alfalfa Silages Prepared with Additives
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McDonald, P.; Henderson, N.; Heron, S. The Biochemistry of Silage, 2nd ed.; Chalcombe Publications: Bucks, UK, 1991; pp. 12, 84–85, 108–111, 167. [Google Scholar]
- Muck, R.E.; Nadeau, E.; Mcallister, T.A.; Contrerasgovea, F.E.; Santos, M.C.; Kung, L. Silage review: Recent advances and future uses of silage additives. J. Dairy Sci. 2018, 101, 3980–4000. [Google Scholar] [CrossRef] [PubMed]
- Muck, R. Recent advances in silage microbiology. Agric. Food Sci. 2013, 22, 3–15. [Google Scholar] [CrossRef]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; Del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lindow, S.E.; Brandl, M.T. Microbiology of the Phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; Coda, R.; De Angelis, M.; Gobbetti, M. Exploitation of vegetables and fruits through lactic acid fermentation. Food Microbiol. 2013, 33, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kung Jr, L. Silage Fermentation & Additives. Available online: https://www.researchgate.net/profile/Limin_Kung/publication/267421247_SILAGE_FERMENTATION_ADDITIVES/links/55ccc4ab08aecae56cc1c3b4/SILAGE-FERMENTATION-ADDITIVES.pdf?origin=publication_detail (accessed on 10 December 2021).
- Zhang, Q.; Yu, Z.; Wang, X. Isolating and evaluating lactic acid bacteria strains with or without sucrose for effectiveness of silage fermentation. Grassl. Sci. 2015, 61, 167–176. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, X.; Wang, C.; He, L.; Zhou, W.; Yang, F.; Zhang, Q. The bacterial community and fermentation quality of mulberry (Morus alba) leaf silage with or without Lactobacillus casei and sucrose. Bioresour. Technol. 2019, 293, 122059. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y. Identification and characterization of Enterococcus species isolated from forage crops and their influence on silage fermentation. J. Dairy Sci. 1999, 82, 2466–2471. [Google Scholar] [CrossRef]
- Tohno, M.; Kobayashi, H.; Nomura, M.; Kitahara, M.; Ohkuma, M.; Uegaki, R.; Cai, Y. Genotypic and phenotypic characterization of lactic acid bacteria isolated from Italian ryegrass silage. Anim. Sci. J. 2012, 83, 111–120. [Google Scholar] [CrossRef]
- Naser, S.M.; Thompson, F.L.; Hoste, B.; Gevers, D.; Dawyndt, P.; Vancanneyt, M.; Swings, J. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 2005, 151, 2141–2150. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, 242–245. [Google Scholar] [CrossRef]
- Kozaki, M.; Uchimura, T.; Okada, S. Experimental Manual for Lactic Acid Bacteria; Asakurasyoten Press: Tokyo, Japan, 1992; pp. 29–72. [Google Scholar]
- Li, D.; Wang, Y.; Zhang, Y.; Lin, Y.; Yang, F. Evaluation of lactic acid bacteria isolated from alfalfa for silage fermentation. Grassl. Sci. 2018, 64, 190–198. [Google Scholar] [CrossRef]
- Yang, J.; Tan, H.; Cai, Y. Characteristics of lactic acid bacteria isolates and their effect on silage fermentation of fruit residues. J. Dairy Sci. 2016, 99, 5325–5334. [Google Scholar] [CrossRef] [PubMed]
- Broderick, G.A.; Kang, J.H. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media1. J. Dairy Sci. 1980, 63, 64–75. [Google Scholar] [CrossRef]
- Muck, R.E. Dry matter level effects on alfalfa silage quality I. Nitrogen transformations. Trans. ASAE 1987, 30, 7–14. [Google Scholar] [CrossRef]
- Licitra, G.; Hernandez, T.M.; Soest, P.J. Standardization of procedures for nitrogen fractionation of ruminant feeds. Anim. Feed Sci. Technol. 1996, 57, 347–358. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis, 15th ed.; Association of Official Analytical Chemistry: Arlington, VA, USA, 1990. [Google Scholar]
- Murphy, R.P. A method for the extraction of plant samples and the determination of total soluble carbohydrates. J. Sci. Food Agric. 1958, 9, 714–717. [Google Scholar] [CrossRef]
- Van Soest, P.V.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.O.; Leveau, J.H.; Marco, M.L. Abundance, diversity and plant-specific adaptations of plant-associated lactic acid bacteria. Environ. Microbiol. Rep. 2020, 12, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Mercier, J.; Lindow, S.E. Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl. Environ. Microbiol. 2000, 66, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Fhoula, I.; Najjari, A.; Turki, Y.; Jaballah, S.; Ouzari, H. Diversity and antimicrobial properties of lactic acid bacteria isolated from rhizosphere of olive trees and desert truffles of tunisia. BioMed Res. Int. 2013, 2, 405708. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Bolsen, K.K.; Brent, B.E.; Fung, D.Y.C. Epiphytic lactic acid bacteria succession during the pre-ensiling and ensiling periods of alfalfa and maize. J. Appl. Bacteriol. 1992, 73, 375–387. [Google Scholar] [CrossRef]
- Liu, D.D.; Gu, C.T. Lactobacillus pingfangensissp. nov., Lactobacillus daoliensissp. nov., Lactobacillus nangangensissp. nov., Lactobacillus daowaiensissp. nov., Lactobacillus dongliensissp. nov., Lactobacillus songbeiensissp. nov. and Lactobacillus kaifaensissp. nov., isolated from traditional Chinese pickle. Int. J. Syst. Evol. Microbiol. 2019, 69, 3237–3247. [Google Scholar]
- Sánchez-Juanes, F.; Teixeira-Martín, V.; González-Buitrago, J.M.; Velázquez, E.; Flores-Félix, J.D. Identification of species and subspecies of lactic acid bacteria present in spanish cheeses type “torta” by MALDI-TOF MS and pheS gene analyses. Microorganisms 2020, 8, 301. [Google Scholar] [CrossRef]
- Naser, S.M.; Dawyndt, P.; Hoste, B.; Gevers, D.; Vandemeulebroecke, K.; Cleenwerck, I.; Vancanneyt, M.; Swings, J. Identification of lactobacilli by pheS and rpoA gene sequence analyses. Int. J. Syst. Evol. Microbiol. 2007, 57, 2777–2789. [Google Scholar] [CrossRef]
- Nel, S.; Davis, S.B.; Endo, A.; Dicks, L.M. Phylogenetic analysis of Leuconostoc and Lactobacillus species isolated from sugarcane processing streams. MicrobiologyOpen 2020, 9, e1065. [Google Scholar] [CrossRef]
- Wang, S.; Dong, Z.; Li, J.; Chen, L.; Shao, T. Effects of storage temperature and combined microbial inoculants on fermentation end products and microbial populations of Italian ryegrass (Lolium multiflorum Lam.) silage. J. Appl. Microbiol. 2018, 125, 1682–1691. [Google Scholar] [CrossRef]
- Xu, D.M.; Ke, W.C.; Zhang, P.; Li, F.H.; Guo, X.S. Characteristics of Pediococcus pentosaceus Q6 isolated from Elymus nutans growing on the Tibetan Plateau and its application for silage preparation at low temperature. J. Appl. Microbiol. 2019, 126, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.; Qin, G.; Tan, Z.; Li, Z.; Wang, Y.; Cai, Y. Natural populations of lactic acid bacteria associated with silage fermentation as determined by phenotype, 16S ribosomal RNA and recA gene analysis. Syst. Appl. Microbiol. 2011, 34, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Kung, L., Jr.; Shaver, R.D.; Grant, R.J.; Schmidt, R.J. Silage review: Interpretation of chemical, microbial, and organoleptic components of silages. J. Dairy Sci. 2018, 101, 4020–4033. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Chevaux, E.; McAllister, T.; Baah, J.; Drouin, P.; Wang, Y. Impact of Pediococcus pentosaceus and Pichia anomala in combination with chitinase on the preservation of high-moisture alfalfa hay. Grass Forage Sci. 2018, 73, 610–621. [Google Scholar] [CrossRef]
- Fusco, V.; Quero, G.M.; Cho, G.S.; Kabisch, J.; Meske, D.; Neve, H.; Bockelmann, W.; Franz, C.M. The genus Weissella: Taxonomy, ecology and biotechnological potential. Front. Microbiol. 2015, 6, 155. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.H.; Kim, K.H.; Chun, B.H.; Ryu, B.H.; Han, N.S.; Jeon, C.O. A proposal of Leuconostoc mesenteroides subsp. jonggajibkimchii subsp. nov. and reclassification of Leuconostoc mesenteroides subsp. suionicum (Gu et al., 2012) as Leuconostoc suionicum sp. nov. based on complete genome sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2225–2230. [Google Scholar] [PubMed]
- Kandler, O.; Schillinger, U.; Weiss, N. Lactobacillus halotolerans sp. nov., nom. rev. and Lactobacillus minor sp. nov., nom. rev. Syst. Appl. Microbiol. 1983, 4, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Ennahar, S.; Cai, Y.; Fujita, Y. Phylogenetic diversity of lactic acid bacteria associated with paddy rice silage as determined by 16S ribosomal DNA analysis. Appl. Environ. Microbiol. 2003, 69, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Blajman, J.E.; Vinderola, G.; Páez, R.B.; Signorini, M.L. The role of homofermentative and heterofermentative lactic acid bacteria for alfalfa silage: A meta-analysis. J. Agric. Sci 2020, 158, 107–118. [Google Scholar] [CrossRef]
- Zheng, M.L.; Niu, D.Z.; Jiang, D.; Zuo, S.S.; Xu, C.C. Dynamics of microbial community during ensiling direct-cut alfalfa with and without LAB inoculant and sugar. J. Appl. Microbiol. 2017, 122, 1456–1470. [Google Scholar] [CrossRef]
- Muck, R.E. Silage microbiology and its control through additives. Rev. Bras. Zootec. 2010, 39, 182–191. [Google Scholar] [CrossRef]
- Oliveira, A.S.; Weinberg, Z.G.; Ogunade, I.M.; Cervantes, A.A.; Arriola, K.G.; Jiang, Y.; Kim, D.; Li, X.; Gonçalves, M.C.M.; Vyas, D.; et al. Meta-analysis of effects of inoculation with homofermentative and facultative heterofermentative lactic acid bacteria on silage fermentation, aerobic stability, and the performance of dairy cows. J. Dairy Sci. 2017, 100, 4587–4603. [Google Scholar] [CrossRef]
- Rooke, J.A.; Kafilzadeh, F. The effect upon fermentation and nutritive value of silages produced after treatment by three different inoculants of lactic acid bacteria applied alone or in combination. Grass Forage Sci. 1994, 49, 324–333. [Google Scholar] [CrossRef]





| Group | Number of Strains | Representative Strain | Cell Form | Fermentation Type | pH in MRS Broth 1 | ||
|---|---|---|---|---|---|---|---|
| 12 h | 24 h | 48 h | |||||
| A | 4 | GI41 | rod | Homo | 4.65 | 4.30 | 4.23 |
| B | 1 | GI4 | rod | Homo | 4.12 | 3.81 | 3.70 |
| C | 1 | GI65 | rod | Homo | 4.20 | 3.98 | 3.95 |
| D | 6 | GI56 | rod | Hetero | 5.50 | 5.09 | 4.80 |
| E | 1 | GI8 | rod | Homo | 3.84 | 3.75 | 3.67 |
| F | 12 | GI19 | rod | Homo | 3.98 | 3.88 | 3.80 |
| G | 1 | GI51 | cocci | Homo | 4.10 | 4.05 | 3.90 |
| H | 12 | GI50 | cocci | Homo | 4.59 | 4.22 | 4.13 |
| I | 1 | GI46 | cocci | Hetero | 5.16 | 4.35 | 4.24 |
| J | 1 | GI21 | cocci | Hetero | 4.66 | 4.29 | 4.17 |
| K | 1 | GI38 | cocci | Hetero | 4.81 | 4.52 | 4.34 |
| L | 2 | GI40 | cocci | Hetero | 4.82 | 4.19 | 4.20 |
| M | 1 | GI62 | cocci | Hetero | 4.81 | 4.21 | 4.19 |
| N | 2 | GI67 | cocci | Hetero | 5.03 | 4.36 | 4.32 |
| O | 3 | GI70 | cocci | Hetero | 4.60 | 4.39 | 4.40 |
| P | 1 | GI52 | cocci | Homo | 5.99 | 4.84 | 4.42 |
| Q | 2 | GI22 | cocci | Homo | 5.27 | 4.35 | 4.32 |
| R | 1 | GI16 | cocci | Homo | 5.24 | 4.91 | 4.32 |
| S | 1 | GI13 | cocci | Homo | 5.04 | 4.91 | 4.50 |
| T | 19 | GI30 | cocci | Homo | 5.09 | 4.54 | 4.43 |
| Item | Raw Alfalfa | Control | GI19 | GI19+GI51 | GI19+S | GI19+GI51+S | SEM | p-Value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| A | L | A × L | ||||||||
| Microbial populations (log10 cfu/g FM) | ||||||||||
| LAB | 3.30 | 7.14 a | 7.64 a | 7.41 a | 4.74 b | 5.19 b | 0.333 | <0.001 | 0.512 | 0.067 |
| Aerobic bacteria | 5.10 | 8.22 a | 7.18 a | 7.52 a | 4.31 b | 5.12 b | 0.422 | <0.001 | 0.059 | 0.403 |
| Coliform bacteria | 4.62 | 3.42 a | 3.14 ab | 2.72 ab | ND | ND | 0.133 | 0.047 | 0.380 | 0.380 |
| Yeast | 3.25 | ND | ND | ND | 4.74 b | 5.32 a | 0.362 | <0.001 | 0.041 | 0.041 |
| Mold | 3.00 | ND | ND | ND | ND | ND | -- | -- | -- | -- |
| Chemical compositions | ||||||||||
| DM (%) | 25.65 | 24.22 ab | 23.14 b | 23.13 b | 25.25 a | 25.40 a | 0.295 | <0.001 | 0.812 | 0.803 |
| CP (% DM) | 17.87 | 16.06 | 17.29 | 17.41 | 17.20 | 17.46 | 0.212 | 0.973 | 0.693 | 0.878 |
| NDF (% DM) | 43.76 | 43.21 | 43.58 | 43.59 | 43.58 | 42.12 | 0.383 | 0.437 | 0.442 | 0.438 |
| ADF (% DM) | 32.68 | 32.21 | 32.29 | 32.04 | 32.63 | 31.03 | 0.316 | 0.647 | 0.229 | 0.365 |
| WSC (% DM) | 5.82 | 1.10 b | 1.08 b | 1.36 b | 2.38 a | 2.16 a | 0.150 | <0.001 | 0.811 | 0.032 |
| Fermentation characteristics | ||||||||||
| PH | -- | 5.69 a | 4.81 b | 4.76 b | 3.91 c | 3.93 c | 0.181 | <0.001 | 0.567 | 0.174 |
| Lactic acid (% DM) | -- | 1.64 c | 3.62 b | 3.87 b | 7.82 a | 7.10 a | 0.650 | <0.001 | 0.380 | 0.092 |
| Acetic acid (% DM) | -- | 2.28 a | 2.22 a | 1.28 b | 0.31 c | 0.21 c | 0.252 | <0.001 | 0.004 | 0.012 |
| Propionic acid (% DM) | -- | 0.53 a | 0.04 b | 0.05 b | 0.01 b | 0.01 b | 0.062 | 0.015 | 0.658 | 0.658 |
| Butyric acid (% DM) | -- | 0.77 | ND | ND | ND | ND | -- | -- | -- | -- |
| Non-protein N fractions (NPN) (% TN) | ||||||||||
| NPN | 26.34 | 61.86 | 62.18 | 61.72 | 58.04 | 61.93 | 0.747 | 0.272 | 0.333 | 0.228 |
| NH3-N | -- | 18.28 a | 10.49 b | 7.84 b | 1.22 d | 4.68 c | 1.500 | <0.001 | 0.179 | <0.001 |
| FAA-N | -- | 29.00 | 27.00 | 27.01 | 24.29 | 25.95 | 0.637 | 0.104 | 0.442 | 0.442 |
| Peptide-N | 14.00 b | 24.69 a | 26.87 a | 32.52 a | 31.30 a | 1.959 | 0.039 | 0.851 | 0.512 1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Li, F.; Zhang, C.; Gao, J.; Tao, Y. Genotypic and Phenotypic Characteristics of Lactic Acid Bacteria Associated with Forage Plants in the Native Grassland of Western Inner Mongolia and Their Application for Alfalfa Silage Fermentation. Animals 2024, 14, 1394. https://doi.org/10.3390/ani14101394
Li W, Li F, Zhang C, Gao J, Tao Y. Genotypic and Phenotypic Characteristics of Lactic Acid Bacteria Associated with Forage Plants in the Native Grassland of Western Inner Mongolia and Their Application for Alfalfa Silage Fermentation. Animals. 2024; 14(10):1394. https://doi.org/10.3390/ani14101394
Chicago/Turabian StyleLi, Wenlong, Feng Li, Chen Zhang, Jie Gao, and Ya Tao. 2024. "Genotypic and Phenotypic Characteristics of Lactic Acid Bacteria Associated with Forage Plants in the Native Grassland of Western Inner Mongolia and Their Application for Alfalfa Silage Fermentation" Animals 14, no. 10: 1394. https://doi.org/10.3390/ani14101394




