Arbuscular Mycorrhizal Fungi Improve Lycium barbarum Potassium Uptake by Activating the Expression of LbHAK
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth Substrate and AM Fungal Inoculum
2.2. Clone and Bioformation Analysis of LbHAK
2.3. Heterologous Expression of LbHAK in Yeast
2.4. Split-Root Experiment Analysis
2.5. Overexpression of LbHAK in Tobacco
2.6. Measurement of Plant Biomass, Mycorrhizal Colonization, and Potassium and Phosphorus Concentrations
2.7. Relative Gene Expression Analysis
2.8. Statistical Analysis
3. Results
3.1. Identification and Functional Analysis of LbHAK
3.2. Overexpression of LbHAK Increased Growth and AM Colonization of Tobacco
3.3. Overexpression of LbHAK Increased Tobacco Potassium and Phosphorus Uptake
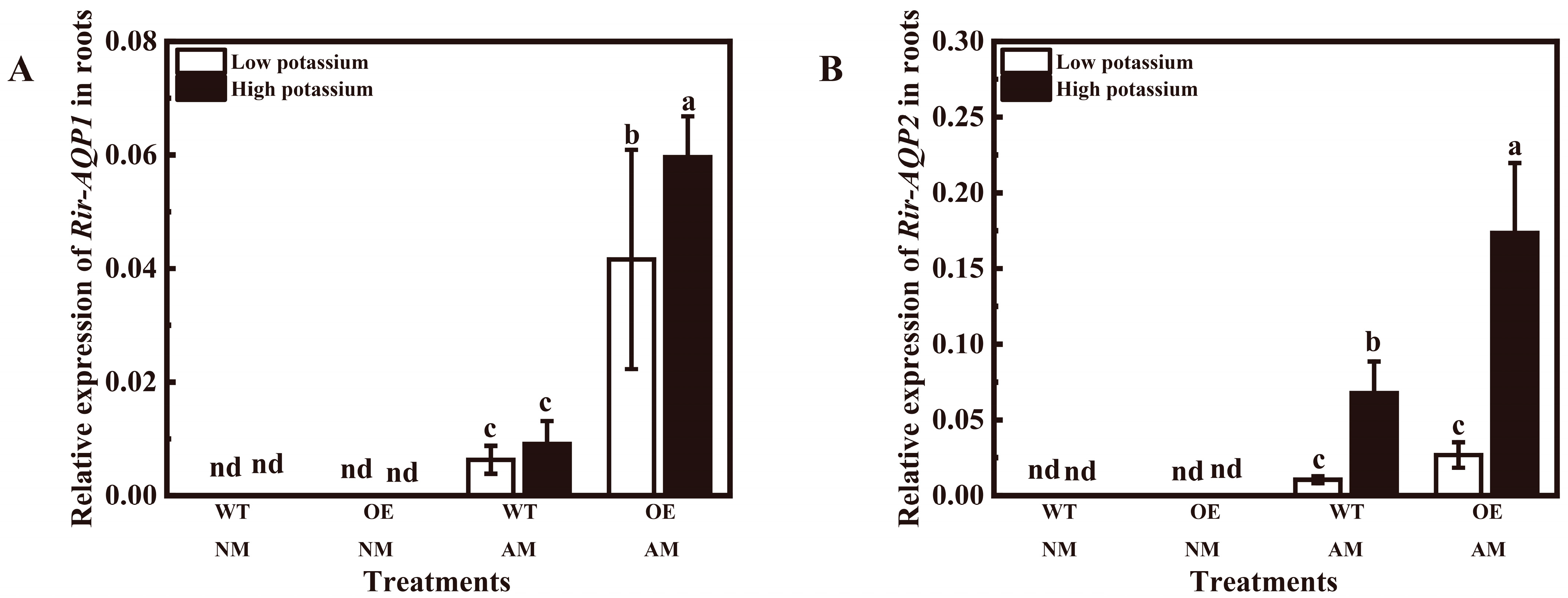
3.4. Overexpression of LbHAK Increased Rir-AQP1 and Rir-AQP2 Expression
3.5. Correlation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, S.E.; Smith, F.A. Roles of Arbuscular Mycorrhizas in Plant Nutrition and Growth: New Paradigms from Cellular to Ecosystem Scales. Annu. Rev. Plant Biol. 2011, 62, 227–250. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.; Qin, C.; Ahanger, M.A.; Raza, S.; Khan, M.I.; Ashraf, M.; Ahmed, N.; Zhang, L. Role of Arbuscular Mycorrhizal Fungi in Plant Growth Regulation: Implications in Abiotic Stress Tolerance. Front. Plant Sci. 2019, 10, 1068. [Google Scholar] [CrossRef] [PubMed]
- Bücking, H.; Kafle, A. Role of Arbuscular Mycorrhizal Fungi in the Nitrogen Uptake of Plants: Current Knowledge and Research Gaps. Agronomy 2015, 5, 587–612. [Google Scholar] [CrossRef]
- Etesami, H.; Jeong, B.R.; Glick, B.R. Contribution of Arbuscular Mycorrhizal Fungi, Phosphate–Solubilizing Bacteria, and Silicon to P Uptake by Plant. Front. Plant Sci. 2021, 12, 699618. [Google Scholar] [CrossRef] [PubMed]
- Javot, H.; Penmetsa, R.V.; Terzaghi, N.; Cook, D.R.; Harrison, M.J. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2007, 104, 1720–1725. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhang, H.; Zhang, X.; Chen, H.; Tang, M. Characterization of six PHT1 members in Lycium barbarum and their response to arbuscular mycorrhiza and water stress. Tree Physiol. 2017, 37, 351–366. [Google Scholar] [PubMed]
- Chen, A.; Hu, J.; Sun, S.; Xu, G. Conservation and divergence of both phosphate- and mycorrhiza-regulated physiological responses and expression patterns of phosphate transporters in Solanaceous species. New Phytol. 2006, 173, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Giovannetti, M.; Tolosano, M.; Volpe, V.; Kopriva, S.; Bonfante, P. Identification and functional characterization of a sulfate transporter induced by both sulfur starvation and mycorrhiza formation in Lotus japonicus. New Phytol. 2014, 204, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, A.; Xie, K.; Yang, X.; Luo, Z.; Chen, J.; Zeng, D.; Ren, Y.; Yang, C.; Wang, L.; et al. Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc. Natl. Acad. Sci. USA 2020, 117, 16649–16659. [Google Scholar] [CrossRef]
- Cheng, K.; Wei, M.; Jin, X.; Tang, M.; Zhang, H. LbAMT3-1, an ammonium transporter induced by arbuscular mycorrhizal in Lycium barbarum, confers tobacco with higher mycorrhizal levels and nutrient uptake. Plant Cell Rep. 2022, 41, 1477–1480. [Google Scholar] [CrossRef]
- Zhang, X.-H.; Lin, A.-J.; Gao, Y.-L.; Reid, R.J.; Wong, M.-H.; Zhu, Y.-G. Arbuscular mycorrhizal colonisation increases copper binding capacity of root cell walls of Oryza sativa L. and reduces copper uptake. Soil Biol. Biochem. 2009, 41, 930–935. [Google Scholar] [CrossRef]
- Han, X.; Zhou, Y.; Li, Y.; Ren, W.; Liu, K.; Zhang, W.; Zhang, H.; Tang, M. LbKAT3 may assist in mycorrhizal potassium uptake, and overexpression of LbKAT3 may promote potassium, phosphorus, and water transport from arbuscular mycorrhizal fungi to the host plant. Front. Plant Sci. 2023, 14, 1161220. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, W.-H. Potassium Transport and Signaling in Higher Plants. Annu. Rev. Plant Biol. 2013, 64, 451–476. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Shankar, A.; Chandran, A.K.N.; Sharma, M.; Jung, K.-H.; Suprasanna, P.; Pandey, G.K. Emerging concepts of potassium homeostasis in plants. J. Exp. Bot. 2019, 71, 608–619. [Google Scholar] [CrossRef] [PubMed]
- Sustr, M.; Soukup, A.; Tylova, E. Potassium in Root Growth and Development. Plants 2019, 8, 435. [Google Scholar] [CrossRef] [PubMed]
- Guillemot, J.; Asensio, V.; Bordron, B.; Nouvellon, Y.; le Maire, G.; Bouillet, J.; Domec, J.; Rojas, J.S.D.; Abreu-Junior, C.H.; Battie-Laclau, P.; et al. Increased hydraulic constraints in Eucalyptus plantations fertilized with potassium. Plant Cell Environ. 2021, 44, 2938–2950. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liao, W. Potassium signaling in plant abiotic responses: Crosstalk with calcium and reactive oxygen species/reactive nitrogen species. Plant Physiol. Biochem. 2022, 173, 110–121. [Google Scholar] [CrossRef]
- Mulet, J.M.; Porcel, R.; Yenush, L. Modulation of potassium transport to increase abiotic stress tolerance in plants. J. Exp. Bot. 2023, 74, 5989–6005. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lyu, C.; Huang, L.; Chen, Q.; Zhuo, W.; Wang, X.; Lu, Y.; Zeng, F.; Lu, L. Physiology and proteomic analysis reveals root, stem and leaf responses to potassium deficiency stress in alligator weed. Sci. Rep. 2019, 9, 17366. [Google Scholar] [CrossRef]
- Battie-Laclau, P.; Laclau, J.-P.; Beri, C.; Mietton, L.; Muniz, M.R.A.; Arenque, B.C.; DE Cassia Piccolo, M.; Jordan-Meille, L.; Bouillet, J.-P.; Nouvellon, Y. Photosynthetic and anatomical responses of Eucalyptus grandis leaves to potassium and sodium supply in a field experiment. Plant Cell Environ. 2014, 37, 70–81. [Google Scholar] [CrossRef]
- Fang, S.; Yang, H.; Wei, G.; Shen, T.; Wan, Z.; Wang, M.; Wang, X.; Wu, Z. Potassium application enhances drought tolerance in sesame by mitigating oxidative damage and regulating osmotic adjustment. Front. Plant Sci. 2022, 13, 1096606. [Google Scholar] [CrossRef]
- Zhu, B.; Xu, Q.; Zou, Y.; Ma, S.; Zhang, X.; Xie, X.; Wang, L. Effect of potassium deficiency on growth, antioxidants, ionome and metabolism in rapeseed under drought stress. Plant Growth Regul. 2019, 90, 455–466. [Google Scholar] [CrossRef]
- Sardans, J.; Peñuelas, J. Potassium Control of Plant Functions: Ecological and Agricultural Implications. Plants 2021, 10, 419. [Google Scholar] [CrossRef]
- Shabala, S.; Pottosin, I. Regulation of potassium transport in plants under hostile conditions: Implications for abiotic and biotic stress tolerance. Physiol. Plant. 2014, 151, 257–279. [Google Scholar] [CrossRef]
- Pettigrew, W.T. Potassium influences on yield and quality production for maize, wheat, soybean and cotton. Physiol. Plant. 2008, 133, 670–681. [Google Scholar] [CrossRef]
- Du, X.-Q.; Wang, F.-L.; Li, H.; Jing, S.; Yu, M.; Li, J.; Wu, W.-H.; Kudla, J.; Wang, Y. The transcription factor MYB59 regulates K+/NO3− translocation in the Arabidopsis response to low K+ stress. Plant Cell 2019, 31, 699–714. [Google Scholar] [CrossRef]
- Rengel, Z.; Damon, P.M. Crops and genotypes differ in efficiency of potassium uptake and use. Physiol. Plant. 2008, 133, 624–636. [Google Scholar] [CrossRef]
- Beier, S.; Marella, N.C.; Yvin, J.-C.; Hosseini, S.A.; von Wirén, N. Silicon mitigates potassium deficiency by enhanced remobilization and modulated potassium transporter regulation. Environ. Exp. Bot. 2022, 198, 104849. [Google Scholar] [CrossRef]
- Garcia, K.; Zimmermann, S.D. The role of mycorrhizal associations in plant potassium nutrition. Front. Plant Sci. 2014, 5, 337. [Google Scholar] [CrossRef]
- Liu, J.; Liu, J.; Liu, J.; Cui, M.; Huang, Y.; Tian, Y.; Chen, A.; Xu, G. The Potassium Transporter SlHAK10 Is Involved in Mycorrhizal Potassium Uptake. Plant Physiol. 2019, 180, 465–479. [Google Scholar] [CrossRef]
- Guether, M.; Balestrini, R.; Hannah, M.; He, J.; Udvardi, M.K.; Bonfante, P. Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol. 2009, 182, 200–212. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, S.; Hu, W.; Xiao, L.; Tang, M. Arbuscular Mycorrhizal Fungus Rhizophagus irregularis Increased Potassium Content and Expression of Genes Encoding Potassium Channels in Lycium barbarum. Front. Plant Sci. 2017, 8, 440. [Google Scholar] [CrossRef]
- Han, X.; Du, X.; Wu, Y.; Wei, M.; Gu, Y.; Aba, X.; Tang, M.; Zhang, H. Foliar-applied potassium improved mycorrhizal Goji (Lycium barbarum L.) growth of the potassium free—Compartment in a compartmented culture system. Sci. Hortic. 2021, 293, 110681. [Google Scholar] [CrossRef]
- Anderson, J.A.; Huprikar, S.S.; Kochian, L.V.; Lucas, W.J.; Gaber, R.F. Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1992, 89, 3736–3740. [Google Scholar] [CrossRef]
- Winston, F.; Dollard, C.; Ricupero-Hovasse, S.L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 1995, 11, 53–55. [Google Scholar] [CrossRef]
- Horie, T.; Brodsky, D.E.; Costa, A.; Kaneko, T.; Lo Schiavo, F.; Katsuhara, M.; Schroeder, J.I. K+ Transport by the OsHKT2; 4 Transporter from Rice with Atypical Na+ Transport Properties and Competition in Permeation of K+ over Mg2+ and Ca2+ Ions. Plant Physiol. 2011, 156, 1493–1507. [Google Scholar] [CrossRef]
- Koske, R.; Gemma, J. A modified procedure for staining roots to detect VA mycorrhizas. Mycol. Res. 1989, 94, 486–488. [Google Scholar] [CrossRef]
- McGONIGLE, T.P.; Miller, M.H.; Evans, D.G.; Fairchild, G.L.; Swan, J.A. A new method which gives an objective measure of colonization of roots by vesicular—Arbuscular mycorrhizal fungi. New Phytol. 1990, 115, 495–501. [Google Scholar] [CrossRef]
- Han, X.; Wang, Y.; Cheng, K.; Zhang, H.; Tang, M. Arbuscular Mycorrhizal Fungus and Exogenous Potassium Application Improved Lycium barbarum Salt Tolerance. J. Plant Growth Regul. 2021, 41, 2980–2991. [Google Scholar] [CrossRef]
- Olsen, S.R.; Sommers, L.E. Phosphorus. In Methods of Soil Analysis Part 2 Chemical and Microbiological Properties, American Society of Agronomy; Page, A.L., Ed.; Soil Science Society of America: Madison, WI, USA, 1982; pp. 403–430. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gong, M.; Zhang, Q.; Cheng, K.; Zhang, H. Symbiosis of arbuscular mycorrhizal fungi and Lycium barbarum L. Prefers NO3− over NH4+. Horticulturae 2023, 9, 637. [Google Scholar] [CrossRef]
- Wu, J.; Ji, J.; Wang, G.; Wu, G.; Diao, J.; Li, Z.; Chen, X.; Chen, Y.; Luo, L. Ectopic expression of the Lycium barbarum β-carotene hydroxylase gene (chyb) enhances drought and salt stress resistance by increasing xanthophyll cycle pool in tobacco. Plant Cell Tissue Organ Cult. (PCTOC) 2015, 121, 559–569. [Google Scholar] [CrossRef]
- Xie, K.; Ren, Y.; Chen, A.; Yang, C.; Zheng, Q.; Chen, J.; Wang, D.; Li, Y.; Hu, S.; Xu, G. Plant nitrogen nutrition: The roles of arbuscular mycorrhizal fungi. J. Plant Physiol. 2021, 269, 153591. [Google Scholar] [CrossRef]
- Jiménez-Leyva, J.A.; Gutiérrez, A.; Orozco, J.A.; Vargas, G.; Esqueda, M.; Gardea, A.; González-Hernández, V.; Sánchez, E.; Muñoz, E. Phenological and ecophysiological responses of Capsicum annuum var. glabriusculum to native arbuscular mycorrhizal fungi and phosphorus availability. Environ. Exp. Bot. 2017, 138, 193–202. [Google Scholar] [CrossRef]
- Garcia, K.; Chasman, D.; Roy, S.; Ané, J.-M. Physiological Responses and Gene Co-Expression Network of Mycorrhizal Roots under K+ Deprivation. Plant Physiol. 2017, 173, 1811–1823. [Google Scholar] [CrossRef]
- Olsson, P.A.; Hammer, E.C.; Pallon, J.; van Aarle, I.M.; Wallander, H. Elemental composition in vesicles of an arbuscular mycorrhizal fungus, as revealed by PIXE analysis. Fungal Biol. 2011, 115, 643–648. [Google Scholar] [CrossRef]
- Olsson, P.l.A.; Hammer, E.C.; Wallander, H.k.; Pallon, J. Phosphorus availability influences elemental uptake in the mycorrhizal fungus Glomus intraradices, as revealed by particle-induced X-ray emission analysis. Appl. Environ. Microbiol. 2008, 74, 4144–4148. [Google Scholar] [CrossRef] [PubMed]
- Bücking, H.; Heyser, W. Elemental composition and function of polyphosphates in ectomycorrhizal fungi—An X-ray microanalytical study. Mycol. Res. 1999, 103, 31–39. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Hijikata, N.; Ohtomo, R.; Handa, Y.; Kawaguchi, M.; Saito, K.; Masuta, C.; Ezawa, T. Aquaporin-mediated long-distance polyphosphate translocation directed towards the host in arbuscular mycorrhizal symbiosis: Application of virus-induced gene silencing. New Phytol. 2016, 211, 1202–1208. [Google Scholar] [CrossRef]
- Reddy, A.; Maley, F. Studies on identifying the catalytic role of Glu-204 in the active site of yeast invertase. J. Biol. Chem. 1996, 271, 13953–13957. [Google Scholar] [CrossRef]
- Liu, L.; Qu, J.; Wang, C.; Liu, M.; Zhang, C.; Zhang, X.; Guo, C.; Wu, C.; Yang, G.; Huang, J.; et al. An efficient genetic transformation system mediated by Rhizobium rhizogenes in fruit trees based on the transgenic hairy root to shoot conversion. Plant Biotechnol. J. 2024. [Google Scholar] [CrossRef]








| Gene Name | GenBank Accession NO. | ORF Length (bp) | Protein Length (bp) | Predicted Molecular Weight (kDa) | Isoelectric Point | Predicted Subcellular Location |
|---|---|---|---|---|---|---|
| LbHAK | MZ416922.1 | 2004 | 668 | 97.35 | 9.07 | Plasma membrane |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Han, X.; Ren, W.; Zhang, H.; Tang, M. Arbuscular Mycorrhizal Fungi Improve Lycium barbarum Potassium Uptake by Activating the Expression of LbHAK. Plants 2024, 13, 1244. https://doi.org/10.3390/plants13091244
Zhang Y, Han X, Ren W, Zhang H, Tang M. Arbuscular Mycorrhizal Fungi Improve Lycium barbarum Potassium Uptake by Activating the Expression of LbHAK. Plants. 2024; 13(9):1244. https://doi.org/10.3390/plants13091244
Chicago/Turabian StyleZhang, Yongxin, Xia Han, Wei Ren, Haoqiang Zhang, and Ming Tang. 2024. "Arbuscular Mycorrhizal Fungi Improve Lycium barbarum Potassium Uptake by Activating the Expression of LbHAK" Plants 13, no. 9: 1244. https://doi.org/10.3390/plants13091244






