Metabolomics in Children Cow’s Milk Protein Allergy: Possible Contribution from a System Biology Approach?
Abstract
:1. Introduction
2. Cow’s Milk Allergenicity and CMA Classification
3. Cow Milk Allergy Diagnosis and Management
4. Origin of Cow Milk Allergy
5. Cow Milk Allergy Primary and Secondary Prevention
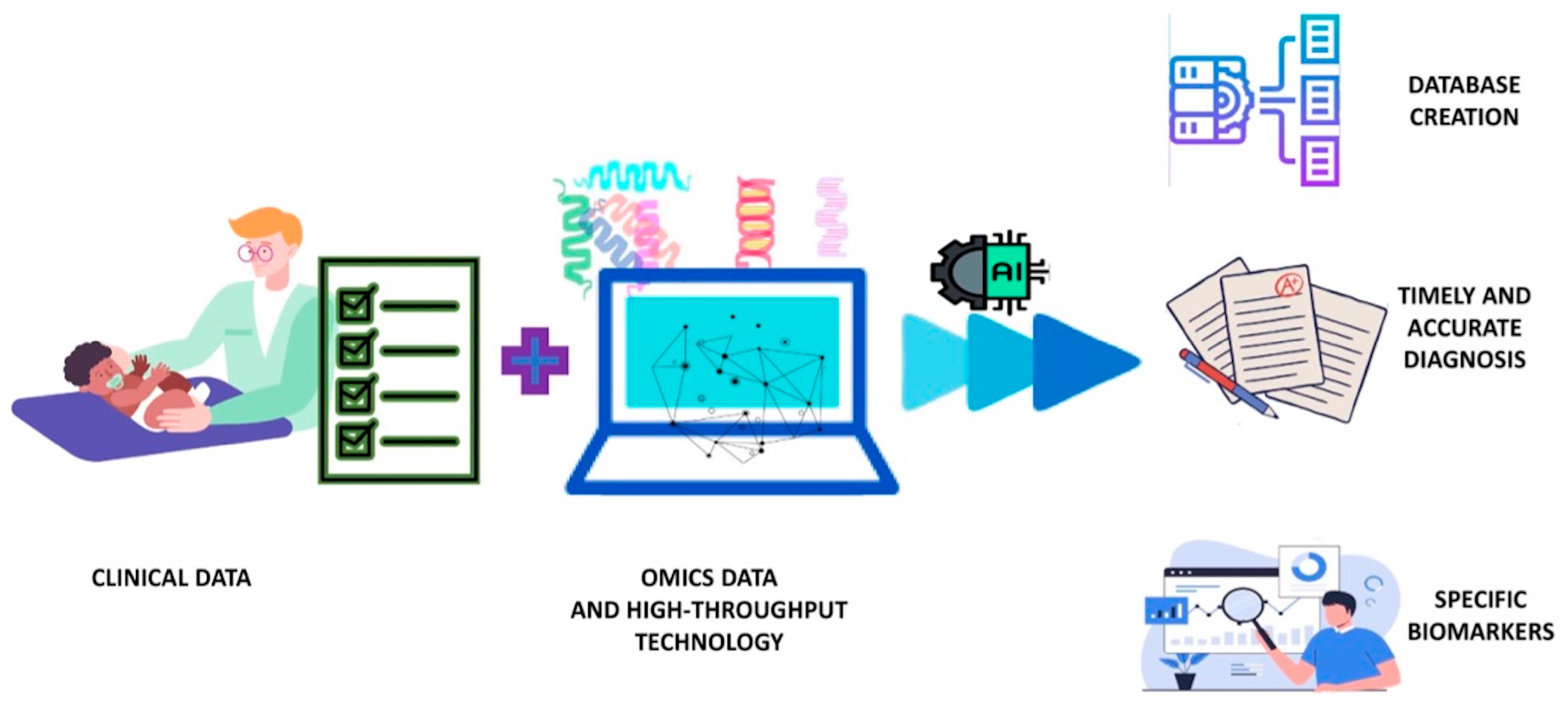
6. Metabolomics and Systems Biology Approach to CMA
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zepeda-Ortega, B.; Goh, A.; Xepapadaki, P.; Sprikkelman, A.; Nicolaou, N.; Hernandez, R.E.H.; Latiff, A.H.A.; Yat, M.T.; Diab, M.; Hussaini, B.A.; et al. Strategies and Future Opportunities for the Prevention, Diagnosis, and Management of Cow Milk Allergy. Front. Immunol. 2021, 12, 608372. [Google Scholar] [CrossRef] [PubMed]
- Flom, J.D.; Sicherer, S.H. Epidemiology of Cow’s Milk Allergy. Nutrients 2019, 11, 1051. [Google Scholar] [CrossRef]
- Meyer, R.; Venter, C.; Bognanni, A.; Szajewska, H.; Shamir, R.; Nowak-Wegrzyn, A.; Fiocchi, A.; Vandenplas, Y.; WAO DRACMA Guideline Group. World Allergy Organization (WAO) Diagnosis and Rationale for Action against Cow’s Milk Allergy (DRACMA) Guideline update—VII—Milk elimination and reintroduction in the diagnostic process of cow’s milk allergy. World Allergy Organ. J. 2023, 16, 100785. [Google Scholar] [CrossRef]
- Calvani, M.; Cardinale, F.; Martelli, A.; Muraro, A.; Pucci, N.; Savino, F.; Zappalà, D.; Panetta, V.; Italian Society of Pediatric Allergy and Immunology Anaphylaxis’ Study Grsoup. Risk factors for severe pediatric food anaphylaxis in Italy. Pediatr. Allergy Immunol. 2011, 22, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Venter, C.; Arshad, S.H. Epidemiology of food allergy. Pediatr. Clin. N. Am. 2011, 58, 327–349. [Google Scholar] [CrossRef] [PubMed]
- Luyt, D.; Ball, H.; Makwana, N.; Green, M.R.; Bravin, K.; Nasser, S.M.; Clark, A.T.; Standards of Care Committee (SOCC) of the British Society for Allergy and Clinical Immunology (BSACI). BSACI guideline for the diagnosis and management of cow’s milk allergy. Clin. Exp. Allergy 2014, 44, 642–672. [Google Scholar] [CrossRef]
- Bajerova, K.; Salvatore, S.; Dupont, C.; Eigenmann, P.; Kuitunen, M.; Meyer, R.; Ribes-Koninckx, C.; Shamir, R.; Szajewska, H.; Vandenplas, Y. The Cow’s Milk-Related Symptom Score (CoMiSS™): A Useful Awareness Tool. Nutrients 2022, 14, 2059. [Google Scholar] [CrossRef]
- Nwaru, B.I.; Hickstein, L.; Panesar, S.S.; Roberts, G.; Muraro, A.; Sheikh, A.; EAACI Food Allergy and Anaphylaxis Guidelines Group. Prevalence of common food allergies in Europe: A systematic review and meta-analysis. Allergy 2014, 69, 992–1007. [Google Scholar] [CrossRef] [PubMed]
- Bavaro, S.L.; De Angelis, E.; Barni, S.; Pilolli, R.; Mori, F.; Novembre, E.M.; Monaci, L. Modulation of Milk Allergenicity by Baking Milk in Foods: A Proteomic Investigation. Nutrients 2019, 11, 1536. [Google Scholar] [CrossRef] [PubMed]
- Salvatore, S.; Agosti, M.; Baldassarre, M.E.; D’Auria, E.; Pensabene, L.; Nosetti, L.; Vandenplas, Y. Cow’s Milk Allergy or Gastroesophageal Reflux Disease-Can We Solve the Dilemma in Infants? Nutrients 2021, 13, 297. [Google Scholar] [CrossRef]
- Cuomo, B.; Indirli, G.C.; Bianchi, A.; Arasi, S.; Caimmi, D.; Dondi, A.; La Grutta, S.; Panetta, V.; Verga, M.C.; Calvani, M. Specific IgE and skin prick tests to diagnose allergy to fresh and baked cow’s milk according to age: A systematic review. Ital. J. Pediatr. 2017, 43, 93. [Google Scholar] [CrossRef]
- D’Auria, E.; Venter, C. Precision medicine in cow’s milk allergy. Curr. Opin. Allergy Clin. Immunol. 2020, 20, 233–241. [Google Scholar] [CrossRef]
- Natale, M.; Bisson, C.; Monti, G.; Peltran, A.; Garoffo, L.P.; Valentini, S.; Fabris, C.; Bertino, E.; Coscia, A.; Conti, A. Cow’s milk allergens identification by two-dimensional immunoblotting and mass spectrometry. Mol. Nutr. Food Res. 2004, 48, 363–369. [Google Scholar] [CrossRef]
- Calvani, M.; Bianchi, A.; Reginelli, C.; Peresso, M.; Testa, A. Oral Food Challenge. Medicina 2019, 55, 651. [Google Scholar] [CrossRef]
- Sampson, H.A.; Ho, D.G. Relationship between food-specific IgE concentrations and the risk of positive food challenges in children and adolescents. J. Allergy Clin. Immunol. 1997, 100, 444–451. [Google Scholar] [CrossRef]
- Schoemaker, A.A.; Sprikkelman, A.B.; Grimshaw, K.E.; Roberts, G.; Grabenhenrich, L.; Rosenfeld, L.; Siegert, S.; Dubakiene, R.; Rudzeviciene, O.; Reche, M.; et al. Incidence and natural history of challenge-proven cow’s milk allergy in European children-EuroPrevall birth cohort. Allergy 2015, 70, 963–972. [Google Scholar] [CrossRef]
- Eigenmann, P.A.; Beyer, K.; Lack, G.; Muraro, A.; Ong, P.Y.; Sicherer, S.H.; Sampson, H.A. Are avoidance diets still warranted in children with atopic dermatitis? Pediatr. Allergy Immunol. 2020, 31, 19–26. [Google Scholar] [CrossRef]
- Dessì, A.; Di Maria, C.; Pintus, R.; Fanos, V.; Bosco, A. Lipidomics and Metabolomics in Infant Atopic Dermatitis: What’s the Correlation with Early Nutrition? Curr. Pediatr. Rev. 2023; online ahead of print. [Google Scholar] [CrossRef]
- Warner, J.O. The early life origins of asthma and related allergic disorders. Arch. Dis. Child. 2004, 89, 97–102. [Google Scholar] [CrossRef]
- Caroli, M.; Vania, A.; Verga, M.C.; Di Mauro, G.; Bergamini, M.; Cuomo, B.; D’Anna, R.; D’Antonio, G.; Dello Iacono, I.; Dessì, A.; et al. Recommendations on Complementary Feeding as a Tool for Prevention of Non-Communicable Diseases (NCDs)-Paper Co-Drafted by the SIPPS, FIMP, SIDOHaD, and SINUPE Joint Working Group. Nutrients 2022, 14, 257. [Google Scholar] [CrossRef] [PubMed]
- Langan, S.M.; Irvine, A.D.; Weidinger, S. Atopic dermatitis. Lancet 2020, 396, 345–360. [Google Scholar] [CrossRef]
- Custovic, A.; Custovic, D.; Kljaić Bukvić, B.; Fontanella, S.; Haider, S. Atopic phenotypes and their implication in the atopic march. Expert. Rev. Clin. Immunol. 2020, 16, 873–881. [Google Scholar] [CrossRef]
- Yang, L.; Fu, J.; Zhou, Y. Research Progress in Atopic March. Front. Immunol. 2020, 11, 1907. [Google Scholar] [CrossRef]
- Aw, M.; Penn, J.; Gauvreau, G.M.; Lima, H.; Sehmi, R. Atopic March: Collegium Internationale Allergologicum Update 2020. Int. Arch. Allergy Immunol. 2020, 181, 1–10. [Google Scholar] [CrossRef]
- Maiello, N.; Comberiati, P.; Giannetti, A.; Ricci, G.; Carello, R.; Galli, E. New Directions in Understanding Atopic March Starting from Atopic Dermatitis. Children 2022, 9, 450. [Google Scholar] [CrossRef]
- Hui-Beckman, J.W.; Goleva, E.; Berdyshev, E.; Leung, D.Y.M. Endotypes of atopic dermatitis and food allergy. J. Allergy Clin. Immunol. 2023, 151, 26–28. [Google Scholar] [CrossRef]
- Leung, D.Y.M.; Calatroni, A.; Zaramela, L.S.; LeBeau, P.K.; Dyjack, N.; Brar, K.; David, G.; Johnson, K.; Leung, S.; Ramirez-Gama, M.; et al. The non lesional skin surface distinguishes atopic dermatitis with food allergy as a unique endotype. Sci. Transl. Med. 2019, 11, eaav2685. [Google Scholar] [CrossRef]
- Papapostolou, N.; Xepapadaki, P.; Gregoriou, S.; Makris, M. Atopic Dermatitis and Food Allergy: A Complex Interplay What We Know and What We Would Like to Learn. J. Clin. Med. 2022, 11, 4232. [Google Scholar] [CrossRef]
- Martin, P.E.; Eckert, J.K.; Koplin, J.J.; Lowe, A.J.; Gurrin, L.C.; Dharmage, S.C.; Vuillermin, P.; Tang, M.L.; Ponsonby, A.L.; Matheson, M.; et al. Which infants with eczema are at risk of food allergy? Results from a population-based cohort. Clin. Exp. Allergy 2015, 45, 255–264. [Google Scholar] [CrossRef]
- Matsumoto, K.; Mori, R.; Miyazaki, C.; Ohya, Y.; Saito, H. Are both early egg introduction and eczema treatment necessary for primary prevention of egg allergy? J. Allergy Clin. Immunol. 2018, 141, 1997–2001.e3. [Google Scholar] [CrossRef]
- Scarpone, R.; Kimkool, P.; Ierodiakonou, D.; Leonardi-Bee, J.; Garcia-Larsen, V.; Perkin, M.R.; Boyle, R.J. Timing of Allergenic Food Introduction and Risk of Immunoglobulin E-Mediated Food Allergy: A Systematic Review and Meta-analysis. JAMA Pediatr. 2023, 177, 489–497. [Google Scholar] [CrossRef]
- Cronin, C.; Ramesh, Y.; De Pieri, C.; Velasco, R.; Trujillo, J. ‘Early Introduction’ of Cow’s Milk for Children with IgE-Mediated Cow’s Milk Protein Allergy: A Review of Current and Emerging Approaches for CMPA Management. Nutrients 2023, 15, 1397. [Google Scholar] [CrossRef]
- Garcia-Larsen, V.; Ierodiakonou, D.; Jarrold, K.; Cunha, S.; Chivinge, J.; Robinson, Z.; Geoghegan, N.; Ruparelia, A.; Devani, P.; Trivella, M.; et al. Diet during pregnancy and infancy and risk of allergic or autoimmune disease: A systematic review and meta-analysis. PLoS Med. 2018, 15, e1002507. [Google Scholar] [CrossRef]
- Demers-Mathieu, V.; Lavangnananda, S. Restricting Cow’s Milk in the Maternal Diet Reduces the Titers of β-Lactoglobulin-Specific IgG Antibodies in Human Milk. Breastfeed. Med. 2022, 17, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Muraro, A.; de Silva, D.; Halken, S.; Worm, M.; Khaleva, E.; Arasi, S.; Dunn-Galvin, A.; Nwaru, B.I.; De Jong, N.W.; Rodríguez Del Río, P.; et al. Managing food allergy: GA2LEN guideline 2022. World Allergy Organ. J. 2022, 15, 100687. [Google Scholar] [CrossRef] [PubMed]
- Vandenplas, Y.; Brough, H.A.; Fiocchi, A.; Miqdady, M.; Munasir, Z.; Salvatore, S.; Thapar, N.; Venter, C.; Vieira, M.C.; Meyer, R. Current Guidelines and Future Strategies for the Management of Cow’s Milk Allergy. J. Asthma Allergy 2021, 14, 1243–1256. [Google Scholar] [CrossRef]
- Järvinen, K.M.; Westfall, J.E.; Seppo, M.S.; James, A.K.; Tsuang, A.J.; Feustel, P.J.; Sampson, H.A.; Berin, C. Role of maternal elimination diets and human milk IgA in the development of cow’s milk allergy in the infants. Clin. Exp. Allergy 2014, 44, 69–78. [Google Scholar] [CrossRef]
- D’Auria, E.; Salvatore, S.; Pozzi, E.; Mantegazza, C.; Sartorio, M.U.A.; Pensabene, L.; Baldassarre, M.E.; Agosti, M.; Vandenplas, Y.; Zuccotti, G. Cow’s Milk Allergy: Immunomodulation by Dietary Intervention. Nutrients 2019, 11, 1399. [Google Scholar] [CrossRef]
- Koukou, Z.; Papadopoulou, E.; Panteris, E.; Papadopoulou, S.; Skordou, A.; Karamaliki, M.; Diamanti, E. The Effect of Breastfeeding on Food Allergies in Newborns and Infants. Children 2023, 10, 1046. [Google Scholar] [CrossRef]
- Doherty, A.M.; Lodge, C.J.; Dharmage, S.C.; Dai, X.; Bode, L.; Lowe, A.J. Human Milk Oligosaccharides and Associations With Immune-Mediated Disease and Infection in Childhood: A Systematic Review. Front. Pediatr. 2018, 6, 91. [Google Scholar] [CrossRef]
- Giannetti, A.; Toschi Vespasiani, G.; Ricci, G.; Miniaci, A.; di Palmo, E.; Pession, A. Cow’s Milk Protein Allergy as a Model of Food Allergies. Nutrients 2021, 13, 1525. [Google Scholar] [CrossRef]
- Fewtrell, M.; Bronsky, J.; Campoy, C.; Domellöf, M.; Embleton, N.; Fidler Mis, N.; Hojsak, I.; Hulst, J.M.; Indrio, F.; Lapillonne, A.; et al. Complementary Feeding: A Position Paper by the European Society for Paediatric Gastroenterology, Hepatology, and Nutrition (ESPGHAN) Committee on Nutrition. J. Pediatr. Gastroenterol. Nutr. 2017, 64, 119–132. [Google Scholar] [CrossRef]
- Perrett, K.P.; Peters, R.L. Emollients for prevention of atopic dermatitis in infancy. Lancet 2020, 395, 923–924. [Google Scholar] [CrossRef] [PubMed]
- Lowe, A.J.; Su, J.C.; Allen, K.J.; Abramson, M.J.; Cranswick, N.; Robertson, C.F.; Forster, D.; Varigos, G.; Hamilton, S.; Kennedy, R.; et al. A randomized trial of a barrier lipid replacement strategy for the prevention of atopic dermatitis and allergic sensitization: The PEBBLES pilot study. Br. J. Dermatol. 2018, 178, e19–e21. [Google Scholar] [CrossRef]
- Lowe, A.; Su, J.; Tang, M.; Lodge, C.J.; Matheson, M.; Allen, K.J.; Varigos, G.; Sasi, A.; Cranswick, N.; Hamilton, S.; et al. PEBBLES study protocol: A randomised controlled trial to prevent atopic dermatitis, food allergy and sensitisation in infants with a family history of allergic disease using a skin barrier improvement strategy. BMJ Open. 2019, 9, e024594. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, M.M.; Phillips, R.; Brown, S.J.; Cro, S.; Cornelius, V.; Carlsen, K.C.L.; Skjerven, H.O.; Rehbinder, E.M.; Lowe, A.J.; Dissanayake, E.; et al. Skin care interventions in infants for preventing eczema and food allergy. Cochrane Database Syst. Rev. 2022, 11, CD013534. [Google Scholar]
- Paparo, L.; Coppola, S.; Nocerino, R.; Pisapia, L.; Picariello, G.; Cortese, M.; Voto, L.; Maglio, M.; Miele, E.; Carucci, L.; et al. How dietary advanced glycation end products could facilitate the occurrence of food allergy. J. Allergy Clin. Immunol. 2024, 153, 742–758. [Google Scholar] [CrossRef] [PubMed]
- Osborn, D.A.; Sinn, J.K.; Jones, L.J. Infant formulas containing hydrolysed protein for prevention of allergic disease. Cochrane Database Syst. Rev. 2018, 10, CD003664. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Nocerino, R.; Terrin, G.; Coruzzo, A.; Cosenza, L.; Leone, L.; Troncone, R. Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy: A randomized trial. J. Allergy Clin. Immunol. 2012, 129, 580–582. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Nocerino, R.; Terrin, G.; Frediani, T.; Lucarelli, S.; Cosenza, L.; Passariello, A.; Leone, L.; Granata, V.; Di Costanzo, M.; et al. Formula selection for management of children with cow’s milk allergy influences the rate of acquisition of tolerance: A prospective multicenter study. J. Pediatr. 2013, 163, 771–777.e1. [Google Scholar]
- Paparo, L.; Nocerino, R.; Bruno, C.; Di Scala, C.; Cosenza, L.; Bedogni, G.; Di Costanzo, M.; Mennini, M.; D’Argenio, V.; Salvatore, F.; et al. Randomized controlled trial on the influence of dietary intervention on epigenetic mechanisms in children with cow’s milk allergy: The EPICMA study. Sci. Rep. 2019, 9, 2828. [Google Scholar] [CrossRef]
- Nowak-Wegrzyn, A.; Czerkies, L.; Reyes, K.; Collins, B.; Heine, R.G. Confirmed Hypoallergenicity of a Novel Whey-Based Extensively Hydrolyzed Infant Formula Containing Two Human Milk Oligosaccharides. Nutrients 2019, 11, 1447. [Google Scholar] [CrossRef]
- Sindher, S.B.; Chin, A.R.; Aghaeepour, N.; Prince, L.; Maecker, H.; Shaw, G.M.; Stevenson, D.K.; Nadeau, K.C.; Snyder, M.; Khatri, P.; et al. Advances and potential of omics studies for understanding the development of food allergy. Front. Allergy 2023, 4, 1149008. [Google Scholar] [CrossRef] [PubMed]
- Ogulur, I.; Pat, Y.; Ardicli, O.; Barletta, E.; Cevhertas, L.; Fernandez-Santamaria, R.; Huang, M.; Bel Imam, M.; Koch, J.; Ma, S.; et al. Advances and highlights in biomarkers of allergic diseases. Allergy 2021, 76, 3659–3686. [Google Scholar] [CrossRef] [PubMed]
- Tedner, S.G.; Asarnoj, A.; Thulin, H.; Westman, M.; Konradsen, J.R.; Nilsson, C. Food allergy and hypersensitivity reactions in children and adults—A review. J. Intern. Med. 2022, 291, 283–302. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, Y.; Zhang, H.; Hu, L.; Liu, J.; Wang, L.; Wang, T.; Zhang, H.; Cong, L.; Wang, Q. Pathogenesis of allergic diseases and implications for therapeutic interventions. Signal Transduct. Target. Ther. 2023, 8, 138. [Google Scholar] [CrossRef]
- Sugita, K.; Akdis, C.A. Recent developments and advances in atopic dermatitis and food allergy. Allergol. Int. 2020, 69, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Cianferoni, A.; Annesi-Maesano, I. Precision medicine in atopic diseases. Curr. Opin. Allergy Clin. Immunol. 2019, 19, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Peng, Y.Q.; Agache, I.; Diamant, Z.; Eiwegger, T.; Fokkens, W.J.; Traidl-Hoffmann, C.; Nadeau, K.; O’Hehir, R.E.; O’Mahony, L.; et al. Biomarkers for diagnosis and prediction of therapy responses in allergic diseases and asthma. Allergy 2020, 75, 3039–3068. [Google Scholar] [CrossRef] [PubMed]
- Muraro, A.; Arasi, S. Biomarkers in Food Allergy. Curr. Allergy Asthma Rep. 2018, 18, 64. [Google Scholar] [CrossRef] [PubMed]
- Muraro, A.; Lemanske, R.F., Jr.; Castells, M.; Torres, M.J.; Khan, D.; Simon, H.U.; Bindslev-Jensen, C.; Burks, W.; Poulsen, L.K.; Sampson, H.A.; et al. Precision medicine in allergic disease-food allergy, drug allergy, and anaphylaxis-PRACTALL document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy, Asthma and Immunology. Allergy 2017, 72, 1006–1021. [Google Scholar] [CrossRef] [PubMed]
- Proper, S.P.; Azouz, N.P.; Mersha, T.B. Achieving Precision Medicine in Allergic Disease: Progress and Challenges. Front. Immunol. 2021, 12, 720746. [Google Scholar] [CrossRef]
- Lee, E.; Lee, S.Y.; Kim, H.B.; Yang, S.I.; Yoon, J.; Suh, D.I.; Oh, H.Y.; Ahn, K.; Kim, K.W.; Shin, Y.H.; et al. Insights from the COCOA birth cohort: The origins of childhood allergic diseases and future perspectives. Allergol. Int. 2024, 73, 3–12. [Google Scholar] [CrossRef]
- Dessì, A.; Cesare Marincola, F.; Masili, A.; Gazzolo, D.; Fanos, V. Clinical metabolomics and nutrition: The new frontier in neonatology and pediatrics. Biomed. Res. Int. 2014, 2014, 981219. [Google Scholar] [CrossRef] [PubMed]
- Bardanzellu, F.; Fanos, V. How could metabolomics change pediatric health? Ital. J. Pediatr. 2020, 46, 37. [Google Scholar] [CrossRef] [PubMed]
- Delles, C.; Husi, H. Systems Biology Approach in Hypertension Research. Methods Mol. Biol. 2017, 1527, 69–79. [Google Scholar] [PubMed]
- Karahalil, B. Overview of Systems Biology and Omics Technologies. Curr. Med. Chem. 2016, 23, 4221–4230. [Google Scholar] [CrossRef]
- Pascal, M.; Perez-Gordo, M.; Caballero, T.; Escribese, M.M.; Lopez Longo, M.N.; Luengo, O.; Manso, L.; Matheu, V.; Seoane, E.; Zamorano, M.; et al. Microbiome and Allergic Diseases. Front. Immunol. 2018, 9, 1584. [Google Scholar] [CrossRef] [PubMed]
- Thompson-Chagoyan, O.C.; Fallani, M.; Maldonado, J.; Vieites, J.M.; Khanna, S.; Edwards, C.; Doré, J.; Gil, A. Faecal microbiota and short-chain fatty acid levels in faeces from infants with cow’s milk protein allergy. Int. Arch. Allergy Immunol. 2011, 156, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Sangwan, N.; Stefka, A.T.; Nocerino, R.; Paparo, L.; Aitoro, R.; Calignano, A.; Khan, A.A.; Gilbert, J.A.; Nagler, C.R. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016, 10, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Seppo, A.E.; Autran, C.A.; Bode, L.; Järvinen, K.M. Human milk oligosaccharides and development of cow’s milk allergy in infants. J. Allergy Clin. Immunol. 2017, 139, 708–711.e5. [Google Scholar] [CrossRef] [PubMed]
- Adel-Patient, K.; Lezmi, G.; Castelli, F.A.; Blanc, S.; Bernard, H.; Soulaines, P.; Dumond, P.; Ah-Leung, S.; Lageix, F.; de Boissieu, D.; et al. Deep analysis of immune response and metabolic signature in children with food protein induced enterocolitis to cow’s milk. Clin. Transl. Allergy 2018, 8, 38. [Google Scholar] [CrossRef]
- Shibata, R.; Itoh, N.; Nakanishi, Y.; Kato, T.; Suda, W.; Nagao, M.; Iwata, T.; Yoshida, H.; Hattori, M.; Fujisawa, T.; et al. Gut microbiota and fecal metabolites in sustained unresponsiveness by oral immunotherapy in school-age children with cow’s milk allergy. Allergol. Int. 2024, 73, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Boulangé, C.L.; Pedersen, H.K.; Martin, F.P.; Siegwald, L.; Pallejà Caro, A.; Eklund, A.C.; Jia, W.; Zhang, H.; Berger, B.; Sprenger, N.; et al. Cinnamon Study Investigator Group. An Extensively Hydrolyzed Formula Supplemented with Two Human Milk Oligosaccharides Modifies the Fecal Microbiome and Metabolome in Infants with Cow’s Milk Protein Allergy. Int. J. Mol. Sci. 2023, 24, 11422. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, E.; Mameli, C.; Piras, C.; Cococcioni, L.; Urbani, A.; Zuccotti, G.V.; Roncada, P. Precision medicine in cow’s milk allergy: Proteomics perspectives from allergens to patients. J. Proteom. 2018, 188, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Wehner, M.R.; Levandoski, K.A.; Kulldorff, M.; Asgari, M.M. Research Techniques Made Simple: An Introduction to Use and Analysis of Big Data in Dermatology. J. Invest. Dermatol. 2017, 137, e153–e158. [Google Scholar] [CrossRef] [PubMed]
- Reel, P.S.; Reel, S.; Pearson, E.; Trucco, E.; Jefferson, E. Using machine learning approaches for multi-omics data analysis: A review. Biotechnol. Adv. 2021, 49, 107739. [Google Scholar] [CrossRef]
- Mori, F.; Barni, S.; Liccioli, G.; Novembre, E. Oral Immunotherapy (OIT): A Personalized Medicine. Medicina 2019, 55, 684. [Google Scholar] [CrossRef] [PubMed]
- Devonshire, A.; Gautam, Y.; Johansson, E.; Mersha, T.B. Multi-omics profiling approach in food allergy. World Allergy Organ. J. 2023, 16, 100777. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Xue, W. IgE-Mediated food allergy: Current diagnostic modalities and novel biomarkers with robust potential. Crit. Rev. Food Sci. Nutr. 2023, 63, 10148–10172. [Google Scholar] [CrossRef]
- Dhondalay, G.K.; Rael, E.; Acharya, S.; Zhang, W.; Sampath, V.; Galli, S.J.; Tibshirani, R.; Boyd, S.D.; Maecker, H.; Nadeau, K.C.; et al. Food allergy and omics. J. Allergy Clin. Immunol. 2018, 141, 20–29. [Google Scholar] [CrossRef]

| Authors/Years | Omics Technologies | Samples | Bio-Specimens | Technique | Results | Clinical Significance |
|---|---|---|---|---|---|---|
| Thompson-Chagoyan et al. [69] 2011 | Metabolomic and microbiomic | 46 CMA infants and 46 healthy controls aged 2–12 months | Fecal samples | GLC Fluorescent in situ hybridization and flow cytometry, using a panel of 10 rRNA targeted group- and species-specific oligonucleotide probes | ↑ proportions of the Clostridium coccoides Group, Atopobium cluster and sum of the proportions of the different bacterial groups in CMA infants ↑ concentrations and percentages of butyric acid and BCSFA in CMA infants | Correlation between gut dysbiosis and CMA although no single species or genus appears to play an essential role Bacterial fermentation’s products could represent biomarkers of the pathology |
| Berni-Canani et al. [70] 2015 | Metabolomic and microbiomic | 19 CMA infants before and 20 healthy controls after treatment with EHCF with (n = 12) and without (n = 7) supplementation with LGG | Fecal samples | GC 16s RNA | Blautia, Roseburia, and Coprococcus were significantly enriched by EHCF and LGG treatment only one genus, Oscillospira, discriminated between infants that became tolerant and those that remained allergic ↑ in fecal butyrate levels in most tolerant infants Blautia and Roseburia exhibited specific strain-level demarcations between tolerant and allergic infants | EHCF + LGG promotes tolerance in infants with CMA, influencing the strain-level bacterial community structure of the infant gut |
| Seppo et al. [71] 2017 | Metabolomic | 41 mothers of non-CMA infants and 39 mothers of CMA infants | Stored breast milk | HPLC | ↓ LNFP III in the mothers with a CMA infant mothers with a non-IgE CMA infant were secretors (2′-FL and LNFP I), those with IgE CMA were not 3 seemingly unrelated HMOs, 6′SL, LSTc, and LNFP III (group a), formed a co-expressed cluster, which, together, significantly correlated with CMA status | The Lewis X antigen (FUT 3) that is present in LNFP III and not the FUT2 is associated with protection against CMA ↑ LNFP III concentrations are not required to prevent CMA, other mechanisms must be involved |
| Adel-Paxtient et al. [72] 2019 | Metabolomic | 9 children with CM-FPIES (3 children initially recruited for IgE-CMA but who experienced negative OFC, IgE resolved) and 12 control subjects (6 IgE) | Plasma samples | LC/MS UHPLC LC/ESI-MS-MS | ↓ concentrations of various fatty acids: alpha-hydrostearic acid, 2-hydroxycaproicacid, myristic acid, palmitic acid, and other unidentified methyl and saturated fatty acids in CM-FPIES infant ↑ concentrations of some amino acids and their derivatives, purine metabolites, or vitamins in CM-FPIES patients vs. IgE-CMA patients but less clearly compared to IgE-resolved ones | Specific metabolomic signature identification for patients with CM-FPIES |
| Shibata et al. [73] 2023 | Metabolomic and microbiomic | 32 school-age children with IgE-mediated CMA who underwent OIT for 13 months | Fecal specimens | MS 16s RNA | ↓ levels of milk and casein-specific IgE and ↑ eigenvalue of Bifidobacterium-dominant module are SU-associated factors and they correlated with other gut environmental modules, especially with Mb-09: Lachnospiraceae and WSM-04: Monosaccharides | Identification of clinical and gut environmental factors associated with SU acquisition in CM-OIT |
| Boulangé et al. [74] 2023 | Metabolomic and microbiomic | 190 non-breastfed infants with CMPA until 12 months of age randomized to receive either the HMOs-supplemented formula (n = 94) or control formula (n = 96) | Fecal specimens | UPLC-MS/M shotgun analysis | HMO intake ↓ the developmental progression toward a mature, adult-type microbiome composition ↑ Bifidobacteria and ↓ Proteobacteria after 1 and 3 months of HMO formula feeding ↓ fecal metabolites derived from the bacterial oxidative catabolism (Ehrlich pathway) of BCAAs and aromatic amino acids (isobutyric acid, isovaleric acid, phenylacetic acid, 3,4-hydroxyphenylacetic acid, and 4-cresol sulfate): ↓ of energy-forming amino acid catabolism bacterial bile acid deconjugation maintained at a stable level over time in the HMO group, while it ↓ in the control group, suggesting an HMO-mediated upregulation of BSH activity no significant differences in acetic acid concentrations between feeding groups, but HMO feeding for 1 month maintained ↑ fecal acetic acid levels compared to baseline levels, while it tended to ↓ in the control group. | Supplementation of a whey-based EHF with 2′-FL and LNnT partially corrected the dysbiosis commonly observed in CMA infants shifting the microbiome composition closer to a pattern typical of breastfed infants |
| Possible Contribution from a Systems Biology Approach to CMA | ||
|---|---|---|
| Purpose | Clinical Relevance | Studies |
| Predictive biomarker research | Effective identification of allergy and reduction of side effects associated with DBPCFC or OFC | Thompson-Chagoyan et al. [66] 2011 Adel-Paxtient et al. [69] 2019 |
| Allergic endotypes classifications | Characterization of different allergic endotypes for personalized therapies | Adel-Paxtient et al. [69] 2019 |
| Pathogenetic mechanisms insight | Clinical and environmental factors associated with SU acquisition during BF or therapy (enriched FM, OIT) Evaluation of persistence of allergy or transient desensitization or SU | Berni-Canani et al., 2015 [67] Seppo et al. [68] 2017 Shibata et al. [70] 2023 Boulangé et al. [71] 2023 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bosco, A.; Altea, V.; Beretta, P.; Cacace, R.; Fanos, V.; Dessì, A. Metabolomics in Children Cow’s Milk Protein Allergy: Possible Contribution from a System Biology Approach? Children 2024, 11, 562. https://doi.org/10.3390/children11050562
Bosco A, Altea V, Beretta P, Cacace R, Fanos V, Dessì A. Metabolomics in Children Cow’s Milk Protein Allergy: Possible Contribution from a System Biology Approach? Children. 2024; 11(5):562. https://doi.org/10.3390/children11050562
Chicago/Turabian StyleBosco, Alice, Veronica Altea, Paola Beretta, Roberto Cacace, Vassilios Fanos, and Angelica Dessì. 2024. "Metabolomics in Children Cow’s Milk Protein Allergy: Possible Contribution from a System Biology Approach?" Children 11, no. 5: 562. https://doi.org/10.3390/children11050562





