Unravelling the Function of the Sesquiterpene Cyclase STC3 in the Lifecycle of Botrytis cinerea
Abstract
:1. Introduction
2. Results
2.1. BcStc3 Is a Well-Conserved Protein
| Gene ID a | Accession Number b | Protein Name | % Identity c | aa | pfam d | Name e | EC Number e | Reaction (IUBMB) e | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bcin12g06390 | XP_024552383.1 | BcStc1/BcBot2 | 100.00 | 399 | PF19086 | presilphiperfolanol synthase | 4.2.3.74 | (2E,6E)-farnesyl diphosphate + H2O = presilphiperfolan-8β-ol + diphosphate | |||||
| Bcin08g02350 | XP_001551948.1 | BcStc2 | 13.06 | 100.00 | 329 | PF06330 | trichodiene synthase | 4.2.3.6 | (2E,6E)-farnesyl diphosphate = trichodiene + diphosphate | ||||
| Bcin13g05830 | XP_024552712.1 | BcStc3 | 14.28 | 15.80 | 100.00 | 411 | PF19086 | aristolochene synthase | 4.2.3.9 | (2E,6E)-farnesyl diphosphate = aristolochene + diphosphate | |||
| Bcin04g03550 | XP_001546971.2 | BcStc4 | 16.54 | 20.66 | 23.84 | 100.00 | 441 | PF19086 | ophiobolin F synthase | 4.2.3.145 | (2E,6E,10E,14E)-geranylfarnesyl diphosphate + H2O = ophiobolin F + diphosphate | ||
| Bcin01g03520 | XP_001550978.1 | BcStc5 | 19.19 | 11.76 | 13.93 | 16.09 | 100.00 | 323 | PF19086 | fusicocca-2,10(14)-diene synthase | 4.2.3.43 | geranylgeranyl diphosphate = fusicocca-2,10(14)-diene +diphosphate | |
| Bcin11g06510 | XP_024551950.1 | BcStc7 | 12.77 | 17.44 | 14.95 | 14.33 | 11.21 | 100.00 | 321 | PF06330 | trichodiene synthase | 4.2.3.6 | (2E,6E)-farnesyl diphosphate = trichodiene + diphosphate |
| BcStc1/ BcBot2 | BcStc2 | BcStc3 | BcStc4 | BcStc5 | BcStc7 | ||||||||
2.2. Expression Analysis of the Sesquiterpene Cyclase Gene Family in B. cinerea
2.3. Co-Regulation of Sesquiterpene Cyclase Genes in B. cinerea
2.4. BcStc3 Is Related to Fungal Growth and Stress Responses
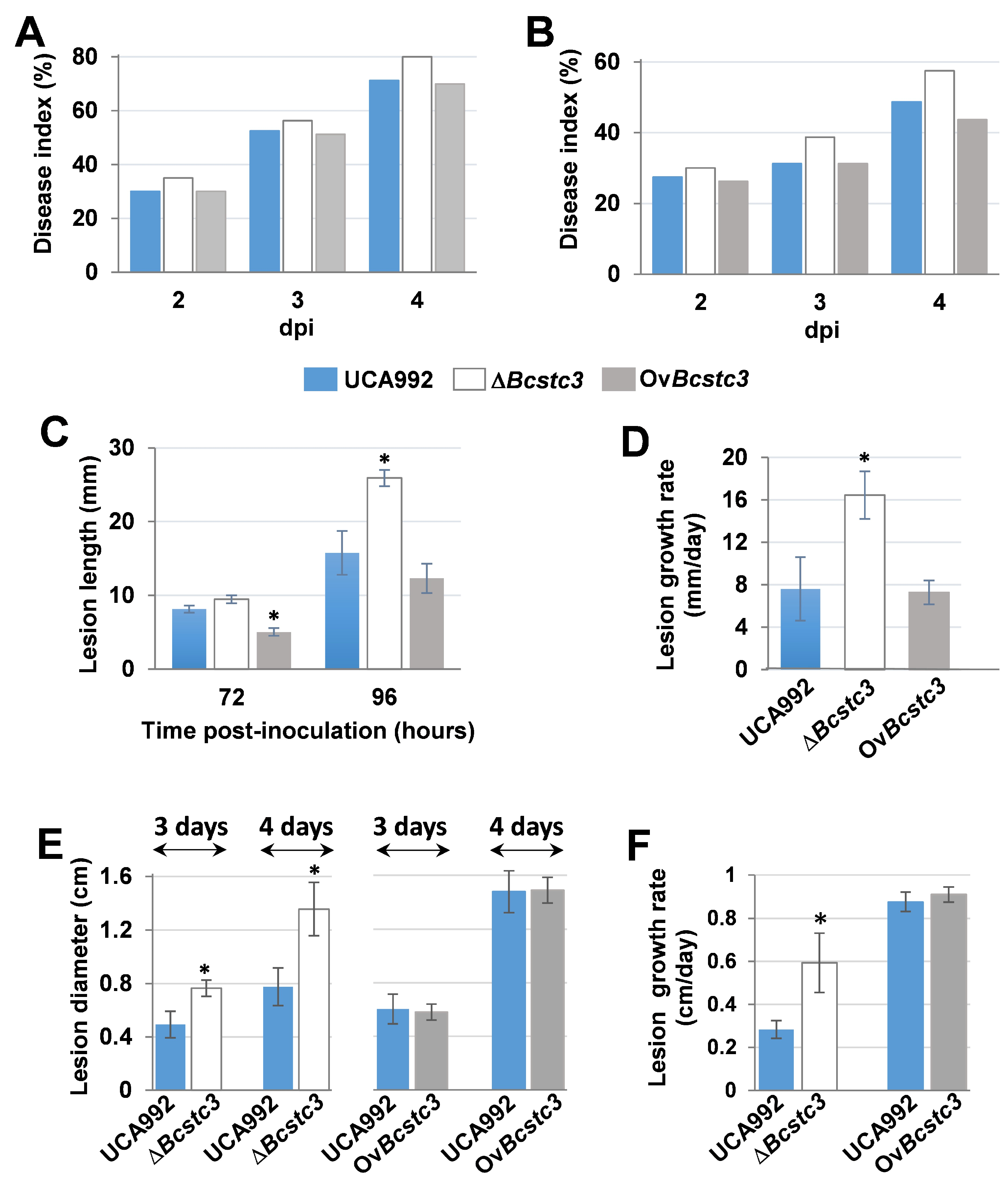
2.5. BcStc3 Is Involved in Virulence
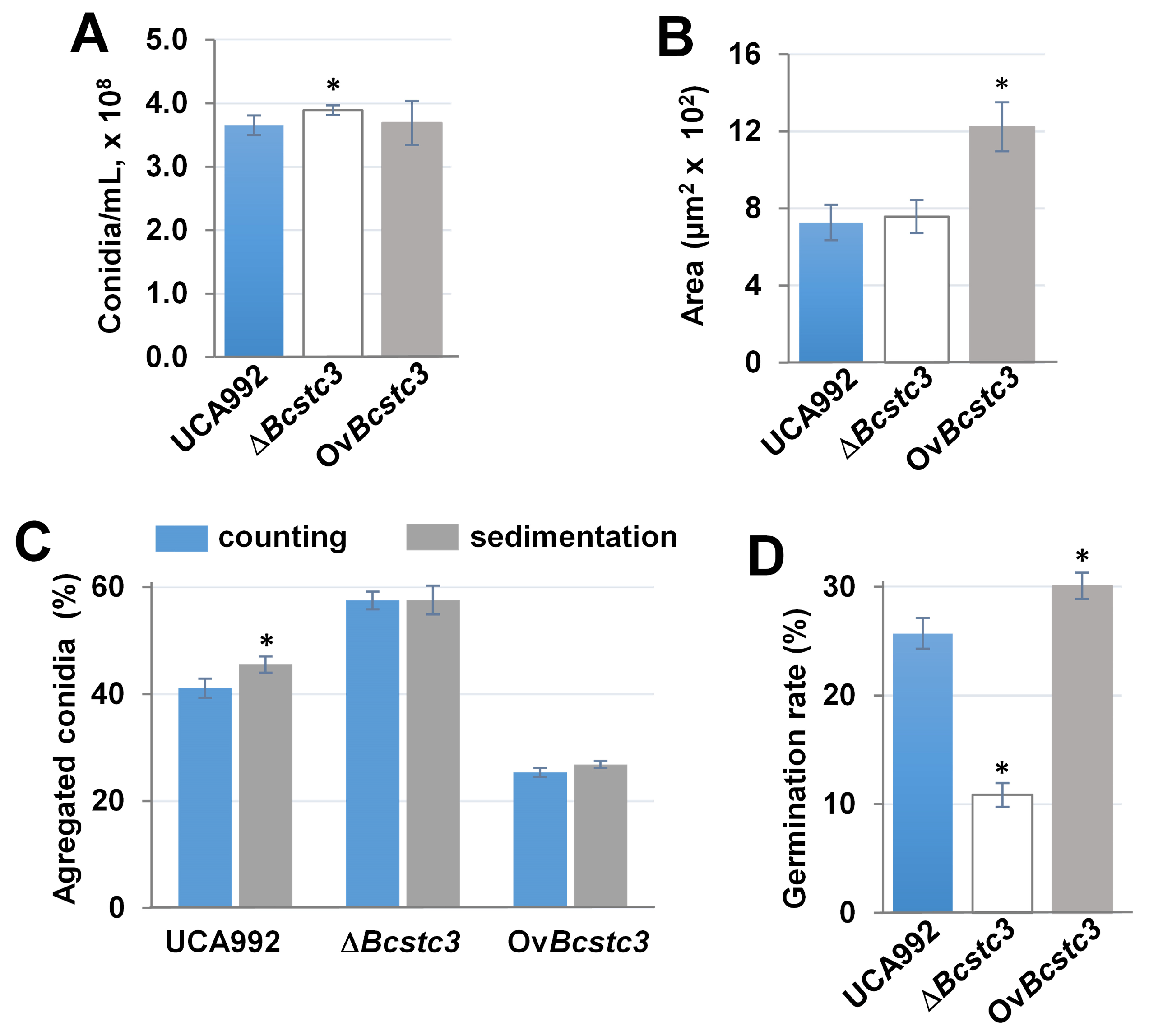
2.6. BcStc3 Is Involved in the Germination and Morphology of Conidia
2.7. Bcstc3 Gene Is Involved in ROS and Infection Cushion Production
2.8. Implication of the BcStc3 on the Secondary Metabolism
3. Discussion
3.1. Bcstc3 Encodes for a Terpene Synthase Family 2, C-Terminal Metal-Binding Domain Protein
3.2. BcStc3 Is a Well-Conserved Protein in Botrytis Genus
3.3. BcStc3 Is a Well-Conserved Protein in Kingdom Fungi
3.4. Bcstc3 Is Differentially Expressed during Fungal Development and Pathogenesis
3.5. BcStc3 Is Involved in Fungal Development and Tolerance to Osmotic and Oxidative Stress
3.6. BcStc3 Is Involved in Conidial Morphogenesis and Infection Cushion Production
3.7. BcStc3 in Involved in Virulence
4. Materials and Methods
4.1. Bioinformatic Analysis
4.2. Organisms, Media and Culture Conditions
4.3. Standard Molecular Methods for Gene Inactivation and Overexpression
4.4. Generation of the ΔBcstc3 and OvBcstc3 Strains
4.5. Quantitative Assessment of Gene Expression via qRT-PCR
4.6. Phenotypic Characterization of Fungal Transformants
4.6.1. Vegetative Growth and Tolerance to Stress Agents
4.6.2. Conidial Production and Germination
4.6.3. Virulence Assays
4.6.4. Reactive Oxygen Species and Infection Cushion Production
4.7. Statistical Analysis
4.8. Metabolomic Characterization of the ΔBcstc3 and Overexpressed OvBcstc3 Mutant Strains
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Minami, A.; Ozaki, T.; Liu, C.; Oikawa, H. Cyclopentane-forming di/sesterterpene synthases: Widely distributed enzymes in bacteria, fungi, and plants. Nat. Prod. Rep. 2018, 35, 1330–1346. [Google Scholar] [CrossRef] [PubMed]
- Christianson, D.W. Unearthing the roots of the terpenome. Curr. Opin. Chem. Biol. 2008, 12, 141–150. [Google Scholar] [CrossRef]
- Tian, S.-H.; Zhang, C.; Zeng, K.-W.; Zhao, M.-B.; Jiang, Y.; Tu, P.-F. Sesquiterpenoids from Artemisia vestita. Phytochemistry 2018, 147, 194–202. [Google Scholar] [CrossRef]
- Zhang, C.; Wen, R.; Ma, X.-L.; Zeng, K.-W.; Xue, Y.; Zhang, P.-M.; Zhao, M.-B.; Jiang, Y.; Liu, G.-Q.; Tu, P.-F. Nitric oxide inhibitory sesquiterpenoids and its dimers from Artemisia freyniana. J. Nat. Prod. 2018, 81, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.-M.; Li, X.-Q.; Chen, C.; Chen, K.; Wang, X.-B.; Gu, Y.-C.; Luo, J.-G.; Kong, L.-Y. Highly oxidized guaianolide sesquiterpenoids with potential anti-inflammatory activity from Chrysanthemum indicum. J. Nat. Prod. 2018, 81, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Yang, B.; Lin, X.; Luo, X.; Pang, X.; Tang, L.; Liu, Y.; Li, X.; Zhou, X. Nitrobenzoyl sesquiterpenoids with cytotoxic activities from a marine-derived Aspergillus ochraceus fungus. J. Nat. Prod. 2018, 81, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Kimani, N.M.; Matasyoh, J.C.; Kaiser, M.; Brun, R.; Schmidt, T.J. Antiprotozoal sesquiterpene lactones and other constituents from Tarchonanthus camphoratus and Schkuhria pinnata. J. Nat. Prod. 2018, 81, 124–130. [Google Scholar] [CrossRef]
- Wang, W.; Liu, Y.; Shi, C.; Pan, L.; Zhang, X.; Zou, J.-J. High energy density renewable fuels based on multicyclic sesquiterpene: Synthesis and performance. Fuel 2022, 318, 123665. [Google Scholar] [CrossRef]
- Whitehead, J.N.; Leferink, N.G.H.; Johannissen, L.O.; Hay, S.; Scrutton, N.S. Decoding catalysis by terpene synthases. ACS Catal. 2023, 13, 12774–12802. [Google Scholar] [CrossRef]
- T, R.; Sharma, D.; Lin, F.; Choong, Y.K.; Lim, C.; Jobichen, C.; Zhang, C. Structural understanding of fungal terpene synthases for the formation of linear or cyclic terpene products. ACS Catal. 2023, 13, 4949–4959. [Google Scholar] [CrossRef]
- González-Hernández, R.A.; Valdez-Cruz, N.A.; Macías-Rubalcava, M.L.; Trujillo-Roldán, M.A. Overview of fungal terpene synthases and their regulation. World J. Microbiol. Biotechnol. 2023, 39, 194. [Google Scholar] [CrossRef] [PubMed]
- Christianson, D.W. Structural biology and chemistry of the terpenoid cyclases. Chem. Rev. 2006, 106, 3412–3442. [Google Scholar] [CrossRef] [PubMed]
- Vavitsas, K.; Fabris, M.; Vickers, C. Terpenoid metabolic engineering in photosynthetic microorganisms. Genes 2018, 9, 520. [Google Scholar] [CrossRef] [PubMed]
- Avalos, M.; Garbeva, P.; Vader, L.; van Wezel, G.P.; Dickschat, J.S.; Ulanova, D. Biosynthesis, evolution and ecology of microbial terpenoids. Nat. Prod. Rep. 2022, 39, 249–272. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Zhang, F.-L.; Feng, T. Sesquiterpenoids specially produced by fungi: Structures, biological activities, chemical and biosynthesis (2015–2020). J. Fungi 2021, 7, 1026. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Xue, M.; Shen, Z.; Jia, X.; Hou, X.; Lai, D.; Zhou, L. Phytotoxic secondary metabolites from fungi. Toxins 2021, 13, 261. [Google Scholar] [CrossRef] [PubMed]
- Shimada, A.; Kusano, M.; Takeuchi, S.; Fujioka, S.; Inokuchi, T.; Kimura, Y. Aspterric acid and 6-Hydroxymellein, inhibitors of pollen development in Arabidopsis thaliana, produced by Aspergillus terreus. Z. Naturforsch. C 2002, 57, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Dalmais, B.; Schumacher, J.; Moraga, J.; Le Pêcheur, P.; Tudzynski, B.; Collado, I.G.; Viaud, M. The Botrytis cinerea phytotoxin botcinic acid requires two polyketide synthases for production and has a redundant role in virulence with botrydial. Mol. Plant Pathol. 2011, 12, 564–579. [Google Scholar] [CrossRef]
- Colmenares, A.J.; Aleu, J.; Durán-Patrón, R.; Collado, I.G.; Hernández-Galán, R. The putative role of botrydial and related metabolites in the infection mechanism of Botrytis cinerea. J. Chem. Ecol. 2002, 28, 997–1005. [Google Scholar] [CrossRef]
- da Silva Ripardo-Filho, H.; Coca Ruíz, V.; Suárez, I.; Moraga, J.; Aleu, J.; Collado, I.G. From genes to molecules, secondary metabolism in Botrytis cinerea: New insights into anamorphic and teleomorphic stages. Plants 2023, 12, 553. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.R.; Gárriz, A.; Marina, M.; Romero, F.M.; Gonzalez, M.E.; Collado, I.G.; Pieckenstain, F.L.; Rubén Rossi, F.; Gárriz, A.; Marina, M.; et al. The sesquiterpene botrydial produced by Botrytis cinerea induces the hypersensitive response on plant tissues and its action is modulated by salicylic acid and jasmonic acid signaling. Mol. Plant-Microbe Interact. 2011, 24, 888–896. [Google Scholar] [CrossRef] [PubMed]
- D’Ambrosio, J.M.; Gonorazky, G.; Sueldo, D.J.; Moraga, J.; Di Palma, A.A.; Lamattina, L.; Collado, I.G.; Laxalt, A.M. The sesquiterpene botrydial from Botrytis cinerea induces phosphatidic acid production in tomato cell suspensions. Planta 2018, 247, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Malmierca, M.G.; Izquierdo-Bueno, I.; Mccormick, S.P.; Cardoza, R.E.; Alexander, N.J.; Moraga, J.; Gomes, E.V.; Proctor, R.H.; Collado, I.G.; Monte, E.; et al. Botrydial and botcinins produced by Botrytis cinerea regulate the expression of Trichoderma arundinaceum genes involved in trichothecene biosynthesis. Mol. Plant Pathol. 2016, 17, 1017–1031. [Google Scholar] [CrossRef] [PubMed]
- Vignatti, P.; Gonzalez, M.E.; Jofré, E.C.; Bolívar-Anillo, H.J.; Moraga, J.; Viaud, M.; Collado, I.G.; Pieckenstain, F.L. Botrydial confers Botrytis cinerea the ability to antagonize soil and phyllospheric bacteria. Fungal Biol. 2020, 124, 54–64. [Google Scholar] [CrossRef]
- Amselem, J.; Cuomo, C.A.; van Kan, J.A.L.L.; Viaud, M.; Benito, E.P.; Couloux, A.; Coutinho, P.M.; de Vries, R.P.; Dyer, P.S.; Fillinger, S.; et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011, 7, e1002230. [Google Scholar] [CrossRef] [PubMed]
- Porquier, A.; Morgant, G.; Moraga, J.; Dalmais, B.; Luyten, I.; Simon, A.; Pradier, J.-M.; Amselem, J.; Collado, I.G.; Viaud, M. The botrydial biosynthetic gene cluster of Botrytis cinerea displays a bipartite genomic structure and is positively regulated by the putative Zn(II)2Cys6 transcription factor BcBot6. Fungal Genet. Biol. 2016, 96, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Pinedo, C.; Wang, C.M.; Pradier, J.M.; Dalmais, B.; Choquer, M.; Le Pêcheur, P.; Morgant, G.; Collado, I.G.; Cane, D.E.; Viaud, M. Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen Botrytis cinerea. ACS Chem. Biol. 2008, 3, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Bueno, I.; González-Rodríguez, V.E.; Simon, A.; Dalmais, B.; Pradier, J.; Le Pêcheur, P.; Mercier, A.; Walker, A.; Garrido, C.; Collado, I.G.; et al. Biosynthesis of abscisic acid in fungi: Identification of a sesquiterpene cyclase as the key enzyme in Botrytis cinerea. Environ. Microbiol. 2018, 20, 2469–2482. [Google Scholar] [CrossRef]
- Suárez, I.; González-Rodríguez, V.E.; Viaud, M.; Garrido, C.; Collado, I.G. Identification of the sesquiterpene cyclase involved in the biosynthesis of (+)-4-Epi-eremophil-9-en-11-ol Derivatives isolated from Botrytis cinerea. ACS Chem. Biol. 2020, 15, 2775–2782. [Google Scholar] [CrossRef]
- Suárez, I.; da Silva Lima, G.; Conti, R.; Pinedo, C.; Moraga, J.; Barúa, J.; de Oliveira, A.L.L.; Aleu, J.; Durán-Patrón, R.; Macías-Sánchez, A.J.; et al. Structural and biosynthetic studies on eremophilenols related to the phytoalexin capsidiol, produced by Botrytis cinerea. Phytochemistry 2018, 154, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Suárez, I.; Pinedo, C.; Aleu, J.; Durán-Patrón, R.; Macías-Sánchez, A.J.; Hernández-Galán, R.; Collado, I.G. The complemented mutant ΔBcstc7, in the STC7 of Botrytis cinerea led to the characterization of 11,12,13-tri-nor-eremophilenols derivatives. Phytochemistry 2022, 193, 113003. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.A.; Barúa, J.E.; Almeida, M.O.; Viaud, M.; Zorrilla, D.; Collado, I.G.; Macías-Sánchez, A.J.; Durán-Patrón, R. Structural and biosynthetic studies of botrycinereic acid, a new cryptic metabolite from the fungus Botrytis cinerea. Bioorg. Chem. 2022, 127, 105979. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Cheng, T.; Wang, A.H.-J. Structure, catalysis, and inhibition mechanism of prenyltransferase. IUBMB Life 2021, 73, 40–63. [Google Scholar] [CrossRef] [PubMed]
- Aaron, J.A.; Christianson, D.W. Trinuclear metal clusters in catalysis by terpenoid synthases. Pure Appl. Chem. 2010, 82, 1585–1597. [Google Scholar] [CrossRef] [PubMed]
- Nosenko, T.; Zimmer, I.; Ghirardo, A.; Köllner, T.G.; Weber, B.; Polle, A.; Rosenkranz, M.; Schnitzler, J.-P. Predicting functions of putative fungal sesquiterpene synthase genes based on multiomics data analysis. Fungal Genet. Biol. 2023, 165, 103779. [Google Scholar] [CrossRef]
- Lou, T.; Li, A.; Xu, H.; Pan, J.; Xing, B.; Wu, R.; Dickschat, J.S.; Yang, D.; Ma, M. Structural insights into three sesquiterpene synthases for the biosynthesis of tricyclic sesquiterpenes and chemical space expansion by structure-based mutagenesis. J. Am. Chem. Soc. 2023, 145, 8474–8485. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- López-Gallego, F.; Wawrzyn, G.; Schmidt-Dannert, C. Selectivity of fungal sesquiterpene synthases: Role of the active site’s H-1α loop in catalysis. Appl. Environ. Microbiol. 2010, 76, 7723–7733. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Pearson, W.R. An Introduction to sequence similarity (“Homology”) searching. Curr. Protoc. Bioinforma. 2013, 42, 3.1.1–3.1.8. [Google Scholar] [CrossRef] [PubMed]
- Choquer, M.; Rascle, C.; Gonçalves, I.R.; de Vallée, A.; Ribot, C.; Loisel, E.; Smilevski, P.; Ferria, J.; Savadogo, M.; Souibgui, E.; et al. The infection cushion of Botrytis cinerea: A fungal ‘weapon’ of plant-biomass destruction. Environ. Microbiol. 2021, 23, 2293–2314. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Koshino, H.; Sakuno, E.; Cutler, H.G.; Nakajima, H. Botcinins E and F and botcinolide from Botrytis cinerea and structural revision of botcinolides. J. Nat. Prod. 2006, 69, 722–725. [Google Scholar] [CrossRef] [PubMed]
- Pinedo, C.; Moraga, J.; Barua, J.; González-Rodríguez, V.E.; Aleu, J.; Durán-Patrón, R.; Macías-Sánchez, A.J.; Hanson, J.R.; Viaud, M.; Hernández-Galán, R.; et al. Chemically induced cryptic sesquiterpenoids and expression of sesquiterpene cyclases in botrytis cinerea revealed new sporogenic (+)-4-epi-eremophil-9-en-11-ols. ACS Chem. Biol. 2016, 11, 1391–1400. [Google Scholar] [CrossRef] [PubMed]
- Nikolaiczyk, V.; Irwan, J.; Nguyen, T.; Fohrer, J.; Elbers, P.; Schrank, P.; Davari, M.D.; Kirschning, A. Rational reprogramming of the sesquiterpene synthase BcBOT2 yields new terpenes with presilphiperfolane skeleton. Catal. Sci. Technol. 2023, 13, 233–244. [Google Scholar] [CrossRef]
- Valero-Jiménez, C.A.; Veloso, J.; Staats, M.; van Kan, J.A.L. Comparative genomics of plant pathogenic Botrytis species with distinct host specificity. BMC Genom. 2019, 20, 203. [Google Scholar] [CrossRef]
- Valero-Jiménez, C.A.; Steentjes, M.B.F.; Slot, J.C.; Shi-Kunne, X.; Scholten, O.E.; van Kan, J.A.L. Dynamics in secondary metabolite gene clusters in otherwise highly syntenic and stable genomes in the fungal genus Botrytis. Genome Biol. Evol. 2020, 12, 2491–2507. [Google Scholar] [CrossRef]
- Garfinkel, A.R. The History of Botrytis taxonomy, the rise of phylogenetics, and implications for species recognition. Phytopathology® 2021, 111, 437–454. [Google Scholar] [CrossRef] [PubMed]
- Hage, H.; Couillaud, J.; Salamov, A.; Loussouarn-Yvon, M.; Durbesson, F.; Ormeño, E.; Grisel, S.; Duquesne, K.; Vincentelli, R.; Grigoriev, I.; et al. An HMM approach expands the landscape of sesquiterpene cyclases across the kingdom fungi. Microb. Genom. 2023, 9, 990. [Google Scholar] [CrossRef] [PubMed]
- Robey, M.T.; Caesar, L.K.; Drott, M.T.; Keller, N.P.; Kelleher, N.L. An interpreted atlas of biosynthetic gene clusters from 1,000 fungal genomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2020230118. [Google Scholar] [CrossRef]
- He, H.; Bian, G.; Herbst-Gervasoni, C.J.; Mori, T.; Shinsky, S.A.; Hou, A.; Mu, X.; Huang, M.; Cheng, S.; Deng, Z.; et al. Discovery of the cryptic function of terpene cyclases as aromatic prenyltransferases. Nat. Commun. 2020, 11, 3958. [Google Scholar] [CrossRef] [PubMed]
- Masi, M.; Meyer, S.; Górecki, M.; Pescitelli, G.; Clement, S.; Cimmino, A.; Evidente, A. Phytotoxic Activity of Metabolites isolated from Rutstroemia sp.n., the causal agent of bleach blonde syndrome on cheatgrass (Bromus tectorum). Molecules 2018, 23, 1734. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Escandón, D.; Tagirdzhanova, G.; Vanderpool, D.; Allen, C.C.G.; Aptroot, A.; Češka, O.; Hawksworth, D.L.; Huereca, A.; Knudsen, K.; Kocourková, J.; et al. Genome-level analyses resolve an ancient lineage of symbiotic ascomycetes. Curr. Biol. 2022, 32, 5209–5218. [Google Scholar] [CrossRef]
- Voglmayr, H.; Fournier, J.; Jaklitsch, W.M. Two new classes of Ascomycota: Xylobotryomycetes and Candelariomycetes. Persoonia-Mol. Phylogeny Evol. Fungi 2019, 42, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Schafhauser, T.; Wibberg, D.; Binder, A.; Rückert, C.; Busche, T.; Wohlleben, W.; Kalinowski, J. Genome assembly and genetic traits of the pleuromutilin-producer Clitopilus passeckerianus DSM1602. J. Fungi 2022, 8, 862. [Google Scholar] [CrossRef]
- Tesei, D. Black fungi research: Out-of-this-world implications. Encyclopedia 2022, 2, 212–229. [Google Scholar] [CrossRef]
- Wu, J.; Yang, X.; Duan, Y.; Wang, P.; Qi, J.; Gao, J.-M.; Liu, C. Biosynthesis of sesquiterpenes in Basidiomycetes: A Review. J. Fungi 2022, 8, 913. [Google Scholar] [CrossRef] [PubMed]
- Kendrick, B. Ainsworth bisbys dictionary of the fungi a review. Mycologist 2003, 17, 17–19. [Google Scholar] [CrossRef]
- Dörner, S.; Rogge, K.; Fricke, J.; Schäfer, T.; Wurlitzer, J.M.; Gressler, M.; Pham, D.N.K.; Manke, D.R.; Chadeayne, A.R.; Hoffmeister, D. Genetic survey of psilocybe natural products. ChemBioChem 2022, 23, e202200249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, X.; Orban, A.; Shukal, S.; Birk, F.; Too, H.-P.; Rühl, M. Agrocybe aegerita serves as a gateway for identifying sesquiterpene biosynthetic enzymes in higher fungi. ACS Chem. Biol. 2020, 15, 1268–1277. [Google Scholar] [CrossRef] [PubMed]
- de Sena Filho, J.G.; Quin, M.B.; Spakowicz, D.J.; Shaw, J.J.; Kucera, K.; Dunican, B.; Strobel, S.A.; Schmidt-Dannert, C. Genome of Diaporthe sp. provides insights into the potential inter-phylum transfer of a fungal sesquiterpenoid biosynthetic pathway. Fungal Biol. 2016, 120, 1050–1063. [Google Scholar] [CrossRef] [PubMed]
- Spatafora, J.W.; Bushley, K.E. Phylogenomics and evolution of secondary metabolism in plant-associated fungi. Curr. Opin. Plant Biol. 2015, 26, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, E.; Seibold, P.S.; Bartram, S.; Trottmann, F.; Haensch, V.G.; Gressler, M.; Chadeayne, A.R.; Hertweck, C.; O’Connor, S.E.; Hoffmeister, D. A “Magic mushroom” multi-product sesquiterpene synthase. ChemBioChem 2023, 24, e202300511. [Google Scholar] [CrossRef] [PubMed]
- Montibus, M.; Pinson-Gadais, L.; Richard-Forget, F.; Barreau, C.; Ponts, N. Coupling of transcriptional response to oxidative stress and secondary metabolism regulation in filamentous fungi. Crit. Rev. Microbiol. 2015, 41, 295–308. [Google Scholar] [CrossRef]
- Umar, A.; Darwish, D.B.E.; Albalwe, F.M. Fungal secondary metabolites and their role in stress management. In Fungal Secondary Metabolites; Elsevier: Amsterdam, The Netherlands, 2024; pp. 15–56. [Google Scholar]
- Overy, D.; Correa, H.; Roullier, C.; Chi, W.-C.; Pang, K.-L.; Rateb, M.; Ebel, R.; Shang, Z.; Capon, R.; Bills, G.; et al. Does osmotic stress affect natural product expression in Fungi? Mar. Drugs 2017, 15, 254. [Google Scholar] [CrossRef]
- Ochiai, N.; Tokai, T.; Nishiuchi, T.; Takahashi-Ando, N.; Fujimura, M.; Kimura, M. Involvement of the osmosensor histidine kinase and osmotic stress-activated protein kinases in the regulation of secondary metabolism in Fusarium graminearum. Biochem. Biophys. Res. Commun. 2007, 363, 639–644. [Google Scholar] [CrossRef]
- Fountain, J.C.; Bajaj, P.; Pandey, M.; Nayak, S.N.; Yang, L.; Kumar, V.; Jayale, A.S.; Chitikineni, A.; Zhuang, W.; Scully, B.T.; et al. Oxidative stress and carbon metabolism influence Aspergillus flavus transcriptome composition and secondary metabolite production. Sci. Rep. 2016, 6, 38747. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, C.; Leng, D.; Yan, P.; Wang, Z.; Zhang, M.; Wu, Z. The non-canonical functions of telomerase reverse transcriptase gene GlTert on regulating fungal growth, oxidative stress, and ganoderic acid biosynthesis in Ganoderma lucidum. Appl. Microbiol. Biotechnol. 2021, 105, 7353–7365. [Google Scholar] [CrossRef]
- Wang, Z.; Lopez-Giraldez, F.; Slot, J.; Yarden, O.; Trail, F.; Townsend, J.P. Secondary metabolism gene clusters exhibit increasingly dynamic and differential expression during asexual growth, conidiation, and sexual development in Neurospora crassa. mSystems 2022, 7, e00232-22. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Yan, J.; Guo, M.; Xu, L.; Hou, L.; Zou, Q. Comparative genome analysis of plant ascomycete fungal pathogens with different lifestyles reveals distinctive virulence strategies. BMC Genom. 2022, 23, 34. [Google Scholar] [CrossRef]
- Udompaisarn, S.; Toopaang, W.; Sae-Ueng, U.; Srisuksam, C.; Wichienchote, N.; Wasuwan, R.; Nahar, N.A.S.; Tanticharoen, M.; Amnuaykanjanasin, A. The polyketide synthase PKS15 has a crucial role in cell wall formation in Beauveria bassiana. Sci. Rep. 2020, 10, 12630. [Google Scholar] [CrossRef] [PubMed]
- Choquer, M.; Fournier, E.; Kunz, C.; Levis, C.; Pradier, J.-M.; Simon, A.; Viaud, M. Botrytis cinerea virulence factors: New insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 2007, 277, 1–10. [Google Scholar] [CrossRef]
- de Vallée, A.; Bally, P.; Bruel, C.; Chandat, L.; Choquer, M.; Dieryckx, C.; Dupuy, J.W.; Kaiser, S.; Latorse, M.-P.; Loisel, E.; et al. A similar secretome disturbance as a hallmark of non-pathogenic Botrytis cinerea ATMT-mutants? Front. Microbiol. 2019, 10, 2829. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; He, Y.; Zhu, P.; Chen, L.; Wang, Y.; Ni, B.; Xu, L. Loss of bcbrn1 and bcpks13 in Botrytis cinerea not only blocks melanization but also increases vegetative growth and virulence. Mol. Plant-Microbe Interact. 2015, 28, 1091–1101. [Google Scholar] [CrossRef]
- Todd, J.N.A.; Carreón-Anguiano, K.G.; Islas-Flores, I.; Canto-Canché, B. Fungal effectoromics: A world in constant evolution. Int. J. Mol. Sci. 2022, 23, 13433. [Google Scholar] [CrossRef]
- Collemare, J.; O’Connell, R.; Lebrun, M. Nonproteinaceous effectors: The terra incognita of plant–fungal interactions. New Phytol. 2019, 223, 590–596. [Google Scholar] [CrossRef]
- Rangel, L.I.; Bolton, M.D. The unsung roles of microbial secondary metabolite effectors in the plant disease cacophony. Curr. Opin. Plant Biol. 2022, 68, 102233. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- González-Rodríguez, V.E.; Garrido, C.; Cantoral, J.M.; Schumacher, J. The F-actin capping protein is required for hyphal growth and full virulence but is dispensable for septum formation in Botrytis cinerea. Fungal Biol. 2016, 120, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, J.W. Fungal nutrition and physiology. Physiol. Plant Pathol. 1985, 26, 120. [Google Scholar] [CrossRef]
- van der Vlugt-Bergmans, C.J.B.; Wagemakers, C.A.M.; van Kan, J.A.L. Cloning and Expression of the cutinase A gene of Botrytis cinerea. Mol. Plant-Microbe Interact. 1997, 10, 21–29. [Google Scholar] [CrossRef]
- Schumacher, J. Tools for Botrytis cinerea: New expression vectors make the gray mold fungus more accessible to cell biology approaches. Fungal Genet. Biol. 2012, 49, 483–497. [Google Scholar] [CrossRef]
- Colot, H.V.; Park, G.; Turner, G.E.; Ringelberg, C.; Crew, C.M.; Litvinkova, L.; Weiss, R.L.; Borkovich, K.A.; Dunlap, J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 2006, 103, 10352–10357. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Cui, K.; He, L.; Zhao, Y.; Mu, W.; Lin, J.; Liu, F. Comparative analysis of Botrytis cinerea in response to the microbial secondary metabolite benzothiazole using iTRAQ-Based quantitative proteomics. Phytopathology® 2021, 111, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Sekulska-Nalewajko, J.; Gocławski, J.; Chojak-Koźniewska, J.; Kuźniak, E. Automated image analysis for quantification of reactive oxygen species in plant leaves. Methods 2016, 109, 114–122. [Google Scholar] [CrossRef] [PubMed]







| Phylum | Sub-Phylum | Super-Class | Fungal Class | Number of Proteins | Number of Organisms |
|---|---|---|---|---|---|
| Ascomycota | Pezizomycotina | Leotiomyceta | Candelariomycetes | 1 | 1 |
| Lecanoromycetes | 9 | 9 | |||
| Eurotiomycetes | 57 | 40 | |||
| Xylobotryomycetes | 1 | 1 | |||
| Dothideomycetes | 40 | 27 | |||
| Sordariomyceta | Leotiomycetes | 11 | 11 | ||
| Sordariomycetes | 34 | 32 | |||
| Basidiomycota | Agaricomycotina | Agaricomycetes | 7 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coca-Ruiz, V.; Suárez, I.; Aleu, J.; Cantoral, J.M.; González, C.; Garrido, C.; Brito, N.; Collado, I.G. Unravelling the Function of the Sesquiterpene Cyclase STC3 in the Lifecycle of Botrytis cinerea. Int. J. Mol. Sci. 2024, 25, 5125. https://doi.org/10.3390/ijms25105125
Coca-Ruiz V, Suárez I, Aleu J, Cantoral JM, González C, Garrido C, Brito N, Collado IG. Unravelling the Function of the Sesquiterpene Cyclase STC3 in the Lifecycle of Botrytis cinerea. International Journal of Molecular Sciences. 2024; 25(10):5125. https://doi.org/10.3390/ijms25105125
Chicago/Turabian StyleCoca-Ruiz, Víctor, Ivonne Suárez, Josefina Aleu, Jesús M. Cantoral, Celedonio González, Carlos Garrido, Nélida Brito, and Isidro G. Collado. 2024. "Unravelling the Function of the Sesquiterpene Cyclase STC3 in the Lifecycle of Botrytis cinerea" International Journal of Molecular Sciences 25, no. 10: 5125. https://doi.org/10.3390/ijms25105125





