Nanoscale Interaction of Endonuclease APE1 with DNA
Abstract
:1. Introduction
2. Results
2.1. DNA Design: Preparation of the Substrate with Two APE1 Sites
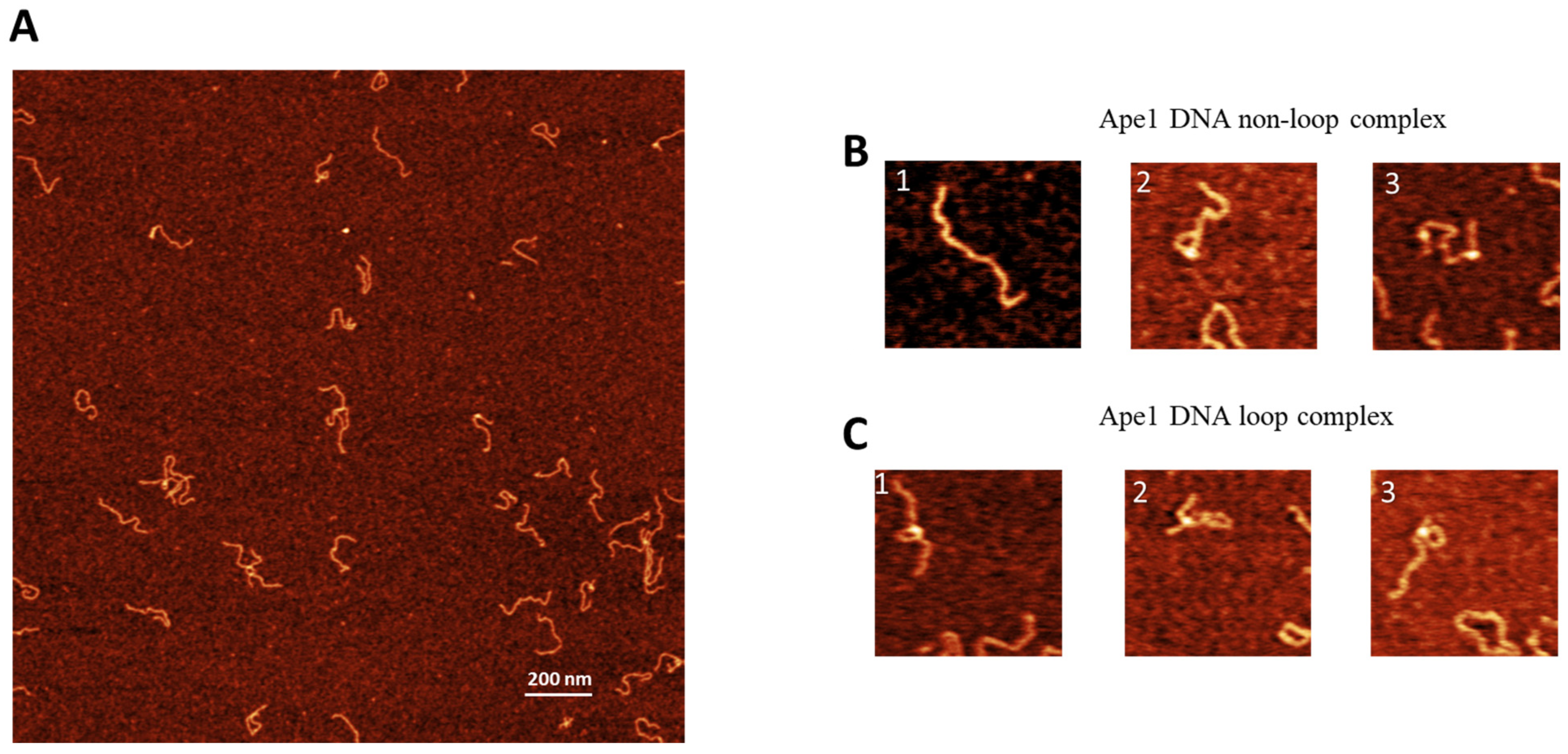
2.2. APE1 Complex Assembly and Loop Size Analysis
2.3. AFM Data Analysis: Positioning of APE1 on DNA
2.4. AFM Data Analysis: Sizes of DNA Loops
2.5. Looped Structures Are Formed by Monomeric APE1
3. Discussion
4. Materials and Methods
4.1. APE1-Protein
4.2. DNA Substrates
4.3. APE1-DNA Synaptosome Assembly
4.4. Atomic Force Microscopy
4.5. Data Analysis
4.6. Height and Volume of APE1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roychoudhury, S.; Pramanik, S.; Harris, H.L.; Tarpley, M.; Sarkar, A.; Spagnol, G.; Sorgen, P.L.; Chowdhury, D.; Band, V.; Klinkebiel, D.; et al. Endogenous oxidized DNA bases and APE1 regulate the formation of G-quadruplex structures in the genome. Proc. Natl. Acad. Sci. USA 2020, 117, 11409–11420. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, S.; Chen, Y.; Song, H.; Khutsishvili, I.; A Marky, L.; Ray, S.; Natarajan, A.; Singh, P.K.; Bhakat, K.K. The human AP-endonuclease 1 (APE1) is a DNA G-quadruplex structure binding protein and regulatesKRASexpression in pancreatic ductal adenocarcinoma cells. Nucleic Acids Res. 2022, 50, 3394–3412. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wilson, D.M. Human Apurinic/Apyrimidinic Endonuclease 1. Antioxid. Redox Signal. 2014, 20, 678–707. [Google Scholar] [CrossRef] [PubMed]
- Bhakat, K.K.; Izumi, T.; Yang, S.; Hazra, T.K.; Mitra, S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003, 22, 6299–6309. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Sarkar, B.; Cholia, R.P.; Gautam, N.; Dhiman, M.; Mantha, A.K. APE1/Ref-1 as an emerging therapeutic target for various human diseases: Phytochemical modulation of its functions. Exp. Mol. Med. 2014, 46, e106. [Google Scholar] [CrossRef]
- Tell, G.; Quadrifoglio, F.; Tiribelli, C.; Kelley, M.R. The Many Functions of APE1/Ref-1: Not Only a DNA Repair Enzyme. Antioxid. Redox Signal. 2009, 11, 601–619. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Dhiman, M.; Tell, G.; Mantha, A.K. A review on protein–protein interaction network of APE1/Ref-1 and its associated biological functions. Cell Biochem. Funct. 2015, 33, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.R.; Limp-Foster, M.; Kelley, M.R. Going APE over ref-1. Mutat. Res. Repair 2000, 461, 83–108. [Google Scholar] [CrossRef]
- Frossi, B.; Antoniali, G.; Yu, K.; Akhtar, N.; Kaplan, M.H.; Kelley, M.R.; Tell, G.; Pucillo, C.E. Endonuclease and redox activities of human apurinic/apyrimidinic endonuclease 1 have distinctive and essential functions in IgA class switch recombination. J. Biol. Chem. 2019, 294, 5198–5207. [Google Scholar] [CrossRef]
- Oliveira, T.T.; Coutinho, L.G.; de Oliveira, L.O.A.; Timoteo, A.R.d.S.; Farias, G.C.; Agnez-Lima, L.F. APE1/Ref-1 Role in Inflammation and Immune Response. Front. Immunol. 2022, 13, 793096. [Google Scholar] [CrossRef]
- Hoitsma, N.M.; Norris, J.; Khoang, T.H.; Kaushik, V.; Chadda, R.; Antony, E.; Hedglin, M.; Freudenthal, B.D. Mechanistic insight into AP-endonuclease 1 cleavage of abasic sites at stalled replication fork mimics. Nucleic Acids Res. 2023, 51, 6738–6753. [Google Scholar] [CrossRef]
- Jaiswal, A.S.; Williamson, E.A.; Srinivasan, G.; Kong, K.; Lomelino, C.L.; McKenna, R.; Walter, C.; Sung, P.; Narayan, S.; Hromas, R. The splicing component ISY1 regulates APE1 in base excision repair. DNA Repair 2019, 86, 102769. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Kim, Y.; Ra, J.S.; Wie, M.W.; Kang, M.-S.; Kang, S.; Myung, K.; Lee, K.-Y. Timely termination of repair DNA synthesis by ATAD5 is important in oxidative DNA damage-induced single-strand break repair. Nucleic Acids Res. 2021, 49, 11746–11764. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Manage, S.A.H.; Burrows, C.J. Binding of AP Endonuclease-1 to G-Quadruplex DNA Depends on the N-Terminal Domain, Mg2+, and Ionic Strength. ACS Bio Med Chem Au 2021, 1, 44–56. [Google Scholar] [CrossRef]
- Dumas, L.; Herviou, P.; Dassi, E.; Cammas, A.; Millevoi, S. G-Quadruplexes in RNA Biology: Recent Advances and Future Directions. Trends Biochem. Sci. 2020, 46, 270–283. [Google Scholar] [CrossRef]
- Pavlova, A.V.; Kubareva, E.A.; Monakhova, M.V.; Zvereva, M.I.; Dolinnaya, N.G. Impact of G-Quadruplexes on the Regulation of Genome Integrity, DNA Damage and Repair. Biomolecules 2021, 11, 1284. [Google Scholar] [CrossRef] [PubMed]
- Miclot, T.; Corbier, C.; Terenzi, A.; Hognon, C.; Grandemange, S.; Barone, G.; Monari, A. Forever Young: Structural Stability of Telomeric Guanine Quadruplexes in the Presence of Oxidative DNA Lesions. Chem.–A Eur. J. 2021, 27, 8865–8874. [Google Scholar] [CrossRef]
- Vascotto, C.; Fantini, D.; Romanello, M.; Cesaratto, L.; Deganuto, M.; Leonardi, A.; Radicella, J.P.; Kelley, M.R.; D’Ambrosio, C.; Scaloni, A.; et al. APE1/Ref-1 Interacts with NPM1 within Nucleoli and Plays a Role in the rRNA Quality Control Process. Mol. Cell. Biol. 2009, 29, 1834–1854. [Google Scholar] [CrossRef]
- Curtis, C.D.; Thorngren, D.L.; Ziegler, Y.S.; Sarkeshik, A.; Yates, J.R.; Nardulli, A.M. Apurinic/Apyrimidinic Endonuclease 1 Alters Estrogen Receptor Activity and Estrogen-Responsive Gene Expression. Mol. Endocrinol. 2009, 23, 1346–1359. [Google Scholar] [CrossRef]
- Lee, Y.R.; Kim, K.M.; Jeon, B.H.; Choi, S. Extracellularly secreted APE1/Ref-1 triggers apoptosis in triple-negative breast cancer cells via RAGE binding, which is mediated through acetylation. Oncotarget 2015, 6, 23383–23398. [Google Scholar] [CrossRef]
- Woo, J.; Park, H.; Sung, S.H.; Moon, B.-I.; Suh, H.; Lim, W. Prognostic Value of Human Apurinic/Apyrimidinic Endonuclease 1 (APE1) Expression in Breast Cancer. PLoS ONE 2014, 9, e99528. [Google Scholar] [CrossRef]
- Bhakat, K.K.; Mantha, A.K.; Mitra, S. Transcriptional Regulatory Functions of Mammalian AP-Endonuclease (APE1/Ref-1), an Essential Multifunctional Protein. Antioxid. Redox Signal. 2009, 11, 621–637. [Google Scholar] [CrossRef]
- Matthews, K.S. DNA looping. Microbiol. Rev. 1992, 56, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Cournac, A.; Plumbridge, J. DNA Looping in Prokaryotes: Experimental and Theoretical Approaches. J. Bacteriol. 2013, 195, 1109–1119. [Google Scholar] [CrossRef] [PubMed]
- Perez, P.J.; Clauvelin, N.; Grosner, M.A.; Colasanti, A.V.; Olson, W.K. What Controls DNA Looping? Int. J. Mol. Sci. 2014, 15, 15090–15108. [Google Scholar] [CrossRef] [PubMed]
- Fogg, J.M.; Judge, A.K.; Stricker, E.; Chan, H.L.; Zechiedrich, L. Supercoiling and looping promote DNA base accessibility and coordination among distant sites. Nat. Commun. 2021, 12, 5683. [Google Scholar] [CrossRef] [PubMed]
- Vilar, J.M.; Saiz, L. DNA looping in gene regulation: From the assembly of macromolecular complexes to the control of transcriptional noise. Curr. Opin. Genet. Dev. 2005, 15, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Lyubchenko, Y.L. Direct AFM visualization of the nanoscale dynamics of biomolecular complexes. J. Phys. D Appl. Phys. 2018, 51, 403001. [Google Scholar] [CrossRef] [PubMed]
- Vemulapalli, S.; Hashemi, M.; Lyubchenko, Y.L. Site-Search Process for Synaptic Protein-DNA Complexes. Int. J. Mol. Sci. 2021, 23, 212. [Google Scholar] [CrossRef]
- Shlyakhtenko, L.S.; Gilmore, J.; Portillo, A.; Tamulaitis, G.; Siksnys, V.; Lyubchenko, Y.L. Direct visualization of the EcoRII− DNA triple synaptic complex by atomic force microscopy. Biochemistry 2007, 46, 11128–11136. [Google Scholar] [CrossRef]
- Lushnikov, A.Y.; Potaman, V.N.; Oussatcheva, E.A.; Sinden, R.R.; Lyubchenko, Y.L. DNA Strand Arrangement within the SfiI-DNA Complex: Atomic Force Microscopy Analysis. Biochemistry 2006, 45, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Shlyakhtenko, L.S.; Lyubchenko, Y.L. High-speed atomic force microscopy directly visualizes conformational dynamics of the HIV Vif protein in complex with three host proteins. J. Biol. Chem. 2020, 295, 11995–12001. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sánchez-Ferrer, A.; Bagnani, M.; Adamcik, J.; Azzari, P.; Hao, J.; Song, A.; Liu, H.; Mezzenga, R. Metal ions confinement defines the architecture of G-quartet, G-quadruplex fibrils and their assembly into nematic tactoids. Proc. Natl. Acad. Sci. USA 2020, 117, 9832–9839. [Google Scholar] [CrossRef]
- Zhou, W.; Lai, R.; Cheng, Y.; Bao, Y.; Miao, W.; Cao, X.; Jia, G.; Li, G.; Li, C. Insights into How NH4+ Ions Enhance the Activity of Dimeric G-Quadruplex/Hemin DNAzyme. ACS Catal. 2023, 13, 4330–4338. [Google Scholar] [CrossRef]
- Tong, X.; Ga, L.; Eerdun, C.; Zhao, R.; Ai, J. Simple Monovalent Metal Ion Logical Order to Regulate the Secondary Conformation of G-Quadruplex. ACS Omega 2022, 7, 39224–39233. [Google Scholar] [CrossRef]
- Schultze, P.; Hud, N.V.; Smith, F.W.; Feigon, J. The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G4T4G4). Nucleic Acids Res. 1999, 27, 3018–3028. [Google Scholar] [CrossRef] [PubMed]
- Marathias, V.M.; Bolton, P.H. Determinants of DNA Quadruplex Structural Type: Sequence and Potassium Binding. Biochemistry 1999, 38, 4355–4364. [Google Scholar] [CrossRef] [PubMed]
- Neaves, K.J.; Huppert, J.L.; Henderson, R.M.; Edwardson, J.M. Direct visualization of G-quadruplexes in DNA using atomic force microscopy. Nucleic Acids Res. 2009, 37, 6269–6275. [Google Scholar] [CrossRef] [PubMed]
- Abdelhady, H.G.; Allen, S.; Davies, M.C.; Roberts, C.J.; Tendler, S.J.B.; Williams, P.M. Direct real-time molecular scale visualisation of the degradation of condensed DNA complexes exposed to DNase I. Nucleic Acids Res. 2003, 31, 4001–4005. [Google Scholar] [CrossRef]
- Kladova, O.A.; Bazlekowa-Karaban, M.; Baconnais, S.; Piétrement, O.; Ishchenko, A.A.; Matkarimov, B.T.; Iakovlev, D.A.; Vasenko, A.; Fedorova, O.S.; Le Cam, E.; et al. The role of the N-terminal domain of human apurinic/apyrimidinic endonuclease 1, APE1, in DNA glycosylase stimulation. DNA Repair 2018, 64, 10–25. [Google Scholar] [CrossRef]
- Pugacheva, E.M.; Kubo, N.; Loukinov, D.; Tajmul, M.; Kang, S.; Kovalchuk, A.L.; Strunnikov, A.V.; Zentner, G.E.; Ren, B.; Lobanenkov, V.V. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc. Natl. Acad. Sci. USA 2020, 117, 2020–2031. [Google Scholar] [CrossRef] [PubMed]
- Mantha, A.K.; Oezguen, N.; Bhakat, K.K.; Izumi, T.; Braun, W.; Mitra, S. Unusual Role of a Cysteine Residue in Substrate Binding and Activity of Human AP-Endonuclease 1. J. Mol. Biol. 2008, 379, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Manage, S.A.H.; Zhu, J.; Fleming, A.M.; Burrows, C.J. Promoters vs. telomeres: AP-endonuclease 1 interactions with abasic sites in G-quadruplex folds depend on topology. RSC Chem. Biol. 2023, 4, 261–270. [Google Scholar] [CrossRef]
- Reshetnikov, R.V.; Kopylov, A.M.; Golovin, A.V. Classification of G-Quadruplex DNA on the Basis of the Quadruplex Twist Angle and Planarity of G-Quartets. Acta Nat. 2010, 2, 72–81. [Google Scholar] [CrossRef]







| Substrate | N = 500 | APE1–DNA Complex (Non-Looped Complexes) | APE1–DNA Complex (Complexes with Loops) |
|---|---|---|---|
| G rich-DNA | Yield [%]. | 31% | 22% |
| Non-G rich DNA | Yield [%]. | 15% | 4% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vemulapalli, S.; Hashemi, M.; Chen, Y.; Pramanik, S.; Bhakat, K.K.; Lyubchenko, Y.L. Nanoscale Interaction of Endonuclease APE1 with DNA. Int. J. Mol. Sci. 2024, 25, 5145. https://doi.org/10.3390/ijms25105145
Vemulapalli S, Hashemi M, Chen Y, Pramanik S, Bhakat KK, Lyubchenko YL. Nanoscale Interaction of Endonuclease APE1 with DNA. International Journal of Molecular Sciences. 2024; 25(10):5145. https://doi.org/10.3390/ijms25105145
Chicago/Turabian StyleVemulapalli, Sridhar, Mohtadin Hashemi, Yingling Chen, Suravi Pramanik, Kishor K. Bhakat, and Yuri L. Lyubchenko. 2024. "Nanoscale Interaction of Endonuclease APE1 with DNA" International Journal of Molecular Sciences 25, no. 10: 5145. https://doi.org/10.3390/ijms25105145






