Immune Rejection Mediated by prf1 and gzmb Affects the Colonization of Fat Greenling (Hexagrammos otakii) Spermatogonia in Heterotransplantation
Abstract
:1. Introduction
2. Results
2.1. Transplantation of Fat Greenling Spermatogonia into Busulfan-Treated Spotted Sea Bass
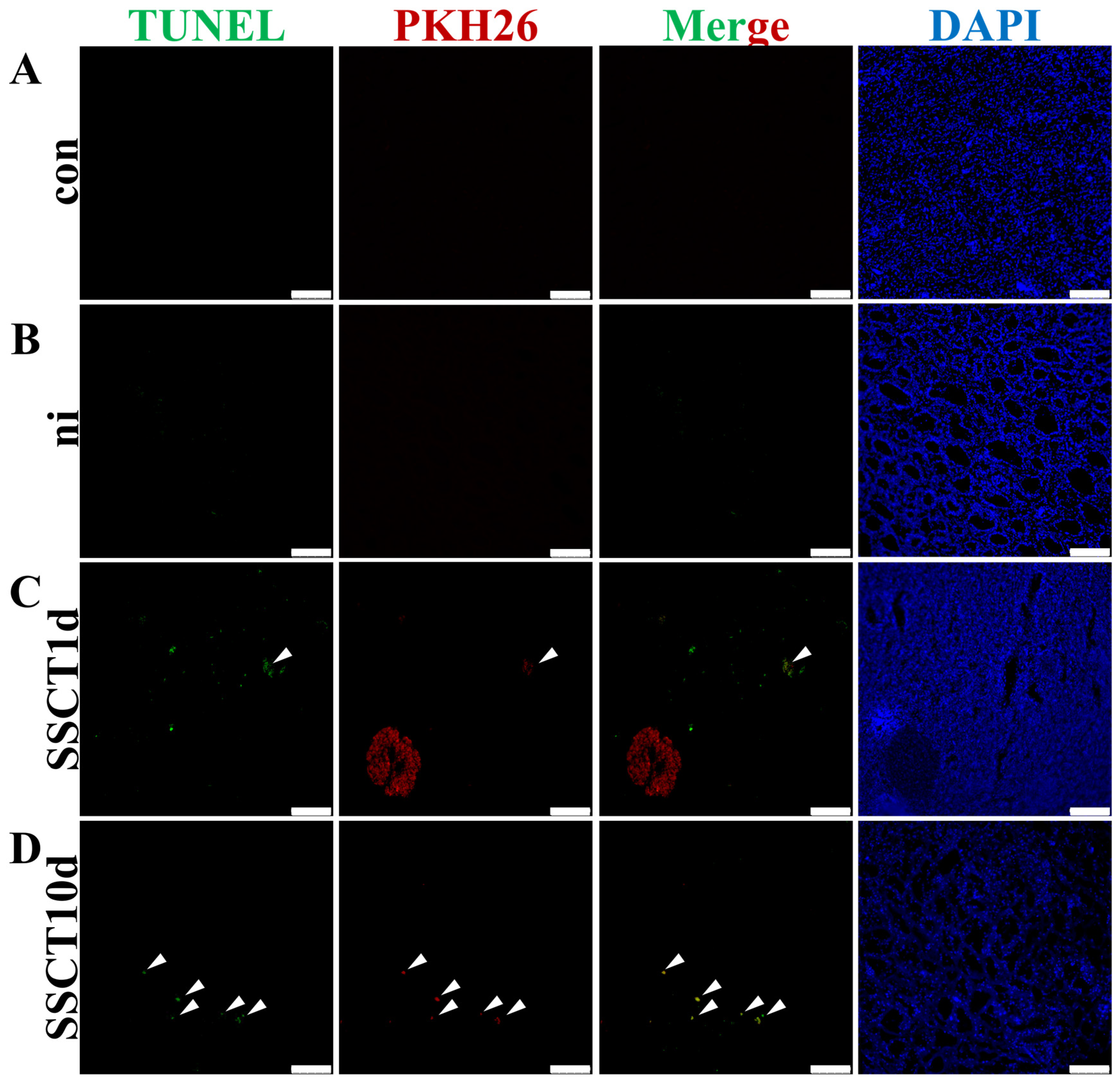
2.2. Apoptosis of Transplanted Spermatogonia
2.3. Differentially Expressed Gene Analysis and Functional Enrichment before and after Transplantation
2.4. Upregulation of Immune-Rejection-Related Genes and Immune Cell Marker Genes after Transplantation
2.5. Analysis of Expression Patterns of prf1 and gzmb Genes and Protein Interaction
2.6. Spotted Sea Bass Testes’ PRF1 and GZMB Induce Apoptosis in Fat Greenling Spermatogonia
3. Discussion
4. Materials and Methods
4.1. Experimental Animal Materials
4.2. Preparation of Sterile Recipients
4.3. Testis Sectioning and Hematoxylin and Eosin (H and E) Staining
4.4. Testicular Tissue RNA Extraction, cDNA Template Synthesis, and qRT-PCR
4.5. Isolation, Purification, and PKH26 Staining of Fat Greenling Spermatogonia
4.6. Experimental Design for Heterotransplantation
4.7. Immunofluorescence of Testicular Tissue
4.8. Detection of Fat Greenling Germ Cells in the Testis of Spotted Sea Bass during the Reproductive Period
4.9. Quality Control and Data Processing
4.10. Differentially Expressed Gene Analysis and Functional Enrichment
4.11. In Situ Hybridization
4.12. Bimolecular Fluorescence Complementation and Immunoprecipitation
4.13. Western Blot
4.14. The Culture and Transfection of Spermatogonia from the Fat Greenling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Majhi, S.K.; Hattori, R.S.; Strussmann, C.A. Transplanted germ cells can colonize the gonads of sexually competent fish and produce functional gametes. Reprod. Fertil. Dev. 2009, 21, 24. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Yoshizaki, G.; Takeuchi, T. Generation of live fry from intraperitoneally transplanted primordial germ cells in rainbow trout. Biol. Reprod. 2003, 69, 1142–1149. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Yoshizaki, G.; Takeuchi, T. Surrogate broodstock produces salmonids. Nature 2004, 430, 629–630. [Google Scholar] [CrossRef]
- Saito, T.; Goto-Kazeto, R.; Arai, K.; Yamaha, E. Xenogenesis in teleost fish through generation of germ-line chimeras by single primordial germ cell transplantation. Biol. Reprod. 2008, 78, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, S.M.; Batlouni, S.R.; Costa, G.M.; Segatelli, T.M.; Quirino, B.R.; Queiroz, B.M.; Kalapothakis, E.; Franca, L.R. A new and fast technique to generate offspring after germ cells transplantation in adult fish: The Nile tilapia (Oreochromis niloticus) model. PLoS ONE 2010, 5, e10740. [Google Scholar] [CrossRef]
- Majhi, S.K.; Hattori, R.S.; Yokota, M.; Watanabe, S.; Strussmann, C.A. Germ cell transplantation using sexually competent fish: An approach for rapid propagation of endangered and valuable germlines. PLoS ONE 2009, 4, e6132. [Google Scholar] [CrossRef] [PubMed]
- Baloch, A.R.; Franek, R.; Tichopad, T.; Fucikova, M.; Rodina, M.; Psenicka, M. Dnd1 knockout in sturgeons by CRISPR/Cas9 generates germ cell free host for surrogate production. Animals 2019, 9, 174. [Google Scholar] [CrossRef]
- Vigoya, A.A.A.; da Costa, D.F.; de Oliveira, M.A.; Butzge, A.J.; Rosa, I.F.; Doretto, L.B.; Martinez, E.R.M.; Digmayer, M.; Nobrega, R.H. Time-efficient germ cell transplantation from goldfish (Carassius auratus) into adult common carp (Cyprinus carpio). Anim. Reprod. 2024, 21, e20230121. [Google Scholar] [CrossRef]
- Goto, R.; Saito, T. A state-of-the-art review of surrogate propagation in fish. Theriogenology 2019, 133, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Franěk, R.; Cheng, Y.; Fučíková, M.; Kašpar, V.; Xie, X.; Shah, M.A.; Linhart, O.; Šauman, I.; Pšenička, M. Who is the best surrogate for germ stem cell transplantation in fish? Aquaculture 2022, 549, 737759. [Google Scholar] [CrossRef]
- Xie, X.; Nobrega, R.; Psenicka, M. Spermatogonial stem cells in fish: Characterization, isolation, enrichment, and recent advances of in vitro culture systems. Biomolecules 2020, 10, 644. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, S.M.S.N.; Aponte, P.M.; Costa, G.M.J.; Campos-Junior, P.H.A.; Segatelli, T.M.; Silva, M.A.; França, L.R. An overview of spermatogonial stem cell physiology, niche and transplantation in fish. Anim. Reprod. 2018, 9, 798–808. [Google Scholar]
- Jones, D.R.E. In utero stem cell transplantation: Two steps forward but one step back? Expert Opin. Biol. Ther. 2001, 1, 205–212. [Google Scholar] [CrossRef] [PubMed]
- El-Awar, N.; Terasaki, P.I.; Cai, J.; Deng, C.-T.; Ozawa, M.; Nguyen, A.; Lias, M.; Conger, N. Epitopes of HLA-A, B, C, DR, DQ, DP and MICA antigens. Clin. Transpl. 2009, 2009, 295–321. [Google Scholar]
- Daar, A.S.; Fuggle, S.V.; Fabre, J.W.; Ting, A.; Morris, P.J. The detailed distribution of HLA-A, B, C antigens in normal human organs. Transplantation 1984, 38, 287–292. [Google Scholar] [CrossRef]
- Sieratzki, J.; Thung, S.N.; Gerber, M.A.; Ferrone, S.; Schaffner, F. Major histocompatibility antigen expression in the liver in acquired immunodeficiency syndrome. Arch. Pathol. Lab. Med. 1987, 111, 1045–1049. [Google Scholar]
- Rosen, H.R. Transplantation immunology: What the clinician needs to know for immunotherapy. Gastroenterology 2008, 134, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F. Male germ cell transplantation: Promise and problems. Reprod. Fertil. Dev. 2001, 13, 609–614. [Google Scholar] [CrossRef]
- Wu, Z.; Tan, X.; Wang, L.; Song, Z.; Wu, Q.; Wang, L.; Yu, Z.; Liu, H.; You, F. Study on artificial induction and early development of gynogenetic fat greenling Hexagrammos otakii. Aquac. Rep. 2022, 22, 100975. [Google Scholar] [CrossRef]
- Wen, H.; Wang, L.; Mou, X.; Chen, C.; Yao, J.; Chen, S. Study on the annual variation of testis development of Hexagrammos otakii jordan and Starks. J. Ocean Univ. China 2007, 37, 581–585. [Google Scholar]
- Liu, J.; Gao, T.; Yokogawa, K.; Zhang, Y. Differential population structuring and demographic history of two closely related fish species, Japanese sea bass (Lateolabrax japonicus) and spotted sea bass (Lateolabrax maculatus) in Northwestern Pacific. Mol. Phylogenet. Evol. 2006, 39, 799–811. [Google Scholar] [CrossRef]
- Yoshizaki, G.; Yazawa, R. Application of surrogate broodstock technology in aquaculture. Fish. Sci. 2019, 85, 429–437. [Google Scholar] [CrossRef]
- Hayashi, M.; Sakuma, D.; Yoshizaki, G. Production of functional sperm by subcutaneous auto-grafting of immature testes in rainbow trout. Mol. Reprod. Dev. 2018, 85, 155–162. [Google Scholar] [CrossRef]
- Nagler, J.J.; Cloud, J.G.; Wheeler, P.A.; Thorgaard, G.H. Testis transplantation in male rainbow trout (Oncorhynchus mykiss). Biol. Reprod. 2001, 64, 644–646. [Google Scholar] [CrossRef]
- Nakanishi, T.; Shibasaki, Y.; Matsuura, Y. T Cells in Fish. Biology 2015, 4, 640–663. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Abdelfattah, N.S.; Blackburn, J.S.; Moore, J.C.; Martinez, S.A.; Moore, F.E.; Lobbardi, R.; Tenente, I.M.; Ignatius, M.S.; Berman, J.N.; et al. Optimized cell transplantation using adult rag2 mutant zebrafish. Nat. Methods 2014, 11, 821–824. [Google Scholar] [CrossRef]
- Ryu, J.H.; Xu, L.; Wong, T.T. Advantages, factors, obstacles, potential solutions, and recent advances of fish germ cell transplantation for aquaculture—A practical review. Animals 2022, 12, 423. [Google Scholar] [CrossRef]
- Ingulli, E. Mechanism of cellular rejection in transplantation. Pediatr. Nephrol. 2010, 25, 61–74. [Google Scholar] [CrossRef]
- Soudais, C.; de Villartay, J.P.; Le Deist, F.; Fischer, A.; Lisowska-Grospierre, B. Independent mutations of the human CD3-ε gene resulting in a T cell receptor/CD3 complex immunodeficiency. Nat. Genet. 1993, 3, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Germain, R.N. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2002, 2, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, Y.; Matsuura, Y.; Toda, H.; Imabayashi, N.; Nishino, T.; Uzumaki, K.; Hatanaka, C.; Yabu, T.; Moritomo, T.; Nakanishi, T. Kinetics of lymphocyte subpopulations in allogeneic grafted scales of ginbuna crucian carp. Dev. Comp. Immunol. 2015, 52, 75–80. [Google Scholar] [CrossRef]
- Trapani, J.A. Target cell apoptosis induced by cytotoxic T cells and natural killer cells involves synergy between the pore-forming protein, perforin, and the serine protease, granzyme B. Aust. N. Z. J. Med. 1995, 25, 793–799. [Google Scholar] [CrossRef]
- Dotiwala, F.; Mulik, S.; Polidoro, R.B.; Ansara, J.A.; Burleigh, B.A.; Walch, M.; Gazzinelli, R.T.; Lieberman, J. Killer lymphocytes use granulysin, perforin and granzymes to kill intracellular parasites. Nat. Med. 2016, 22, 210–216. [Google Scholar] [CrossRef]
- Simon, T.; Opelz, G.; Wiesel, M.; Ott, R.C.; Süsal, C. Serial peripheral blood perforin and granzyme B gene expression measurements for prediction of acute rejection in kidney graft recipients. Am. J. Transplant. 2003, 3, 1121–1127. [Google Scholar] [CrossRef] [PubMed]
- Boivin, W.A.; Cooper, D.M.; Hiebert, P.R.; Granville, D.J. Intracellular versus extracellular granzyme B in immunity and disease: Challenging the dogma. Lab. Investig. 2009, 89, 1195–1220. [Google Scholar] [CrossRef] [PubMed]
- Rousalova, I.; Krepela, E. Granzyme B-induced apoptosis in cancer cells and its regulation. Int. J. Oncol. 2010, 37, 1361–1378. [Google Scholar] [CrossRef]
- Perlmutter, R.M. Translational Regulation of the Lymphocyte-Specific Protein Tyrosine Kinase p56^ lck1. Enzyme 1990, 44, 214–224. [Google Scholar] [CrossRef]
- Stirnweiss, A.; Hartig, R.; Gieseler, S.; Lindquist, J.A.; Reichardt, P.; Philipsen, L.; Simeoni, L.; Poltorak, M.; Merten, C.; Zuschratter, W. T cell activation results in conformational changes in the Src family kinase Lck to induce its activation. Sci. Signal. 2013, 6, ra13. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Aguilar, L.M.; Vico-Barranco, I.; Arbulo-Echevarria, M.M.; Aguado, E. A story of kinases and adaptors: The role of LCK, ZAP-70 and LAT in switch panel governing T-cell development and activation. Biology 2023, 12, 1163. [Google Scholar] [CrossRef]
- Chen, J.; Moore, A.; Ringshausen, I. ZAP-70 shapes the immune microenvironment in B cell malignancies. Front. Oncol. 2020, 10, 595832. [Google Scholar] [CrossRef]
- Hauber, I.; Gulle, H.; Wolf, H.M.; Maris, M.; Eggenbauer, H.; Eibl, M.M. Molecular characterization of major histocompatibility complex class II gene expression and demonstration of antigen-specific T cell response indicate a new phenotype in class II-deficient patients. J. Exp. Med. 1995, 181, 1411–1423. [Google Scholar] [CrossRef]
- Jeon, G.H.; Lee, S.H.; Cheon, Y.P.; Choi, D. Blood-testis barrier and sperm delayed in the cauda epididymis of the reproductively regressed Syrian hamsters. Dev. Reprod. 2021, 25, 1–14. [Google Scholar] [CrossRef]
- Wanjari, U.R.; Gopalakrishnan, A.V. A review on immunological aspects in male reproduction: An immune cells and cytokines. J. Reprod. Immunol. 2023, 158, 103984. [Google Scholar] [CrossRef]
- Valdez, B.C.; Murray, D.; Nieto, Y.; Li, Y.; Wang, G.; Champlin, R.E.; Andersson, B.S. Synergistic cytotoxicity of the DNA alkylating agent busulfan, nucleoside analogs and suberoylanilide hydroxamic acid in lymphoma cell lines. Leuk. Lymphoma 2012, 53, 973–981. [Google Scholar] [CrossRef]
- Kraynak, A.R.; Storer, R.D.; Jensen, R.D.; Kloss, M.W.; Soper, K.A.; Clair, J.H.; DeLuca, J.G.; Nichols, W.W.; Eydelloth, R.S. Extent and persistence of streptozotocin-induced DNA damage and cell proliferation in rat kidney as determined by in vivo alkaline elution and BrdUrd labeling assays. Toxicol. Appl. Pharmacol. 1995, 135, 279–286. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhao, J.; Qi, H.; Li, X.; Zhang, S.; Song, D.W.; Yu, C.; Gao, J. Accelerated ovarian aging in mice by treatment of busulfan and cyclophosphamide. J. Zhejiang Univ. Sci. B 2013, 14, 318–324. [Google Scholar] [CrossRef]
- Huguley, C.M.; Grizzle, J.; Rundles, R.W.; Bell, W.N.; Corley, C.C.; Frommeyer, W.B.; Greenberg, B.G.; Hammack, W.; Herion, J.C.; James, G.W.; et al. Comparison of 6-mercaptopurine and busulfan in chronic granulocytic leukemia. Blood 1963, 21, 89–101. [Google Scholar] [CrossRef]
- Bucci, L.R.; Meistrich, M.L. Effects of busulfan on murine spermatogenesis: Cytotoxicity, sterility, sperm abnormalities, and dominant lethal mutations. Mutat. Res. 1987, 176, 259–268. [Google Scholar] [CrossRef]
- Xu, J.; You, F.; Wu, X.; Zhang, P.; Lin, Y.; Jiang, H.; Zheng, C. Induction of triploidy in large yellow crocker Pseudosciaena crocea (Richardson, 1846): Effects of pressure shocks and growth performance in the first rearing year. Aquac. Res. 2008, 39, 1369–1376. [Google Scholar] [CrossRef]
- Jiang, S.; Xu, Y.; Fan, Y.; Hu, Y.; Zhang, Q.; Su, W. Busulfan impairs blood–testis barrier and spermatogenesis by increasing noncollagenous 1 domain peptide via matrix metalloproteinase 9. Andrology 2022, 10, 377–391. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, M.; Wang, Y.; Xu, J.; Ma, C.; Tang, Y.; Luo, Q.; Zhang, H.; Xu, F. Endoplasmic reticulum stress promotes blood-testis barrier impairment in mice with busulfan-induced oligospermia through PERK-eIF2α signaling pathway. Toxicology 2022, 473, 153193. [Google Scholar] [CrossRef]
- Hillebrand, G.; Castro, L.A.; Illner, W.D.; Schleibner, S.; Land, W.; Gurland, H.J. Combined immunosuppression (cyclosporin, azathioprine, methylprednisolone) in patients at immunological risk after kidney transplantation. Z. Urol. Nephrol. 1985, 78, 667–671. [Google Scholar]
- Ghoneim, M.A.; Sobh, M.A.; Shokeir, A.A.; Bakr, M.A.; El-Sherif, A.K.; Fouda, M.A. Prospective randomized study of azathioprine versus cyclosporin in live-donor kidney transplantation. Am. J. Nephrol. 1993, 13, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Herrid, M.; Davey, R.J.; Hutton, K.; Colditz, I.G.; Hill, J.R. A comparison of methods for preparing enriched populations of bovine spermatogonia. Reprod. Fertil. Dev. 2009, 21, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Miao, G.; Shao, C.; Tian, Y.; Liao, X. Isolation and characterization of polymorphic microsatellite loci from fat greenling (Hexagrammos otakii). Conserv. Genet. 2009, 10, 1429–1431. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Dewey, C.N.; Bo, L. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Chen, Y.; Li, R.; Men, Y.; Yan, K.; Li, Z.; Cai, W.; He, Y.; Qi, J. Immune Rejection Mediated by prf1 and gzmb Affects the Colonization of Fat Greenling (Hexagrammos otakii) Spermatogonia in Heterotransplantation. Int. J. Mol. Sci. 2024, 25, 5157. https://doi.org/10.3390/ijms25105157
Zhao X, Chen Y, Li R, Men Y, Yan K, Li Z, Cai W, He Y, Qi J. Immune Rejection Mediated by prf1 and gzmb Affects the Colonization of Fat Greenling (Hexagrammos otakii) Spermatogonia in Heterotransplantation. International Journal of Molecular Sciences. 2024; 25(10):5157. https://doi.org/10.3390/ijms25105157
Chicago/Turabian StyleZhao, Xi, Ying Chen, Rui Li, Yu Men, Kai Yan, Zibin Li, Wenxiu Cai, Yan He, and Jie Qi. 2024. "Immune Rejection Mediated by prf1 and gzmb Affects the Colonization of Fat Greenling (Hexagrammos otakii) Spermatogonia in Heterotransplantation" International Journal of Molecular Sciences 25, no. 10: 5157. https://doi.org/10.3390/ijms25105157





