ccdC Regulates Biofilm Dispersal in Bacillus velezensis FZB42
Abstract
:1. Introduction
2. Results
2.1. The ccdC Gene Regulates Biofilm Development in B. velezensis FZB42
2.2. ccdC Is More Highly Expressed in Mature Biofilms Than in Early Biofilms
2.3. ccdC Is Essential for Biofilm Dispersal
2.4. ccdC Regulates Sporulation and Motility
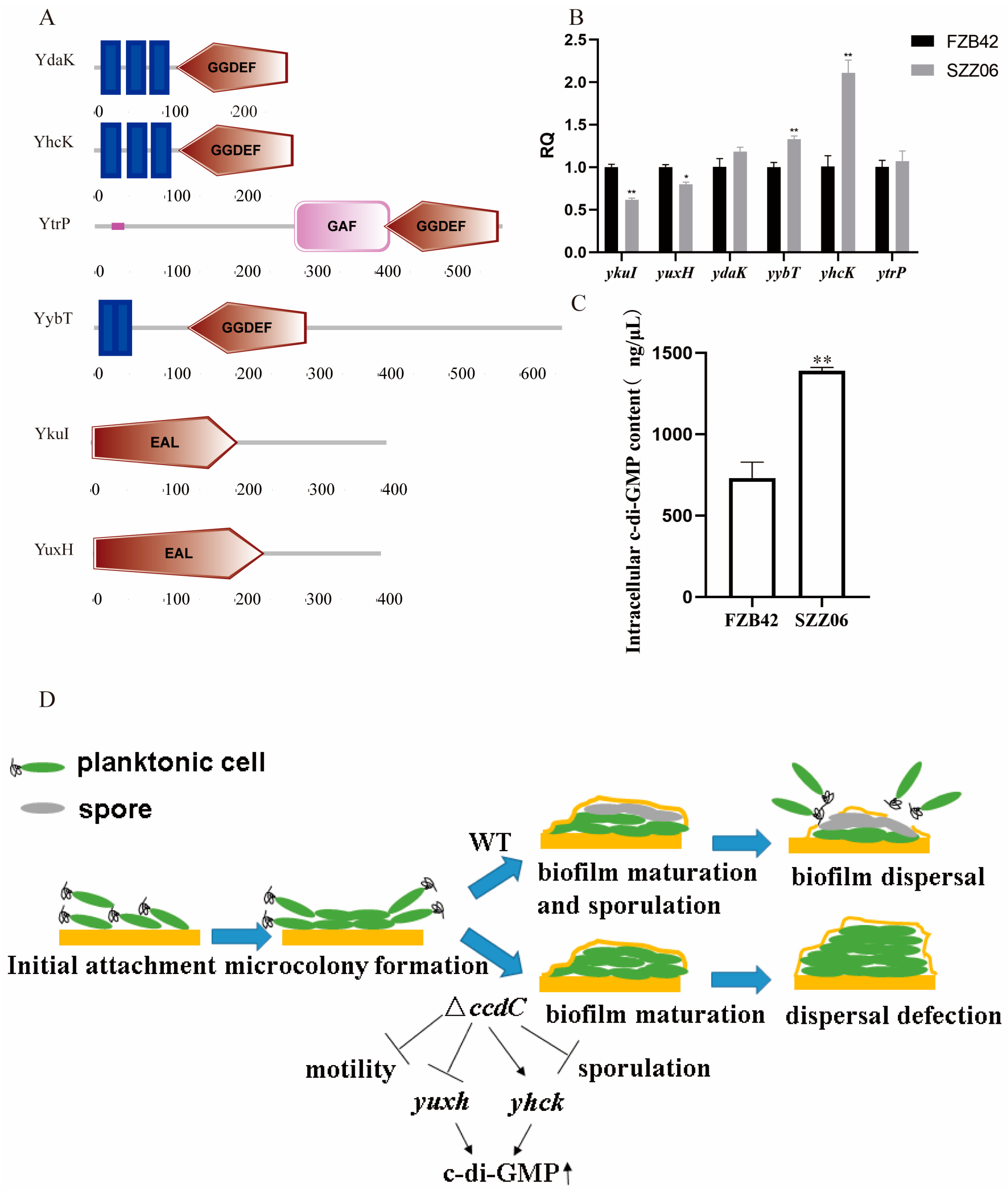
2.5. ccdC Modulates c-di-GMP Levels to Regulate Biofilm Dispersal
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Transposon Mutant Library
4.3. Mutant Status Verification
4.4. Biofilm Development Assay
4.5. Biofilm Dispersal Assay
4.6. Swarming Motility Assay
4.7. Spore Formation Assay
4.8. Construction of the ccdC Knockout Strain and Complement Strain
4.9. Quantitative Real-Time Polymerase Chain Reaction (qRT—PCR)
4.10. Intracellular c-di-GMP Measurement Assay
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the Natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Trejo, M.; Douarche, C.; Bailleux, V.; Poulard, C.; Mariot, S.; Regeard, C.; Raspaud, E. Elasticity and wrinkled morphology of Bacillus subtilis pellicles. Proc. Natl. Acad. Sci. USA 2013, 110, 2011–2016. [Google Scholar] [CrossRef] [PubMed]
- Fröls, S. Archaeal biofilms: Widespread and complex. Biochem. Soc. Trans. 2013, 41, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Fleming, D.; Rumbaugh, K.P. Approaches to Dispersing Medical Biofilms. Microorganisms 2017, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The First 25 Years of a Universal Bacterial Second Messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef] [PubMed]
- Petrova, O.E.; Sauer, K. Escaping the biofilm in more than one way: Desorption, detachment or dispersion. Curr. Opin. Microbiol. 2016, 30, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Chua, S.L.; Tan, S.Y.-Y.; Rybtke, M.T.; Chen, Y.; Rice, S.A.; Kjelleberg, S.; Tolker-Nielsen, T.; Yang, L.; Givskov, M. Bis-(3′-5′)-Cyclic Dimeric GMP Regulates Antimicrobial Peptide Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 2066–2075. [Google Scholar] [CrossRef] [PubMed]
- Chua, S.L.; Liu, Y.; Yam, J.K.H.; Chen, Y.; Vejborg, R.M.; Tan, B.G.C.; Kjelleberg, S.; Tolker-Nielsen, T.; Givskov, M.; Yang, L. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 2014, 5, 4462. [Google Scholar] [CrossRef]
- Marks, L.R.; Davidson, B.A.; Knight, P.R.; Hakansson, A.P. Interkingdom signaling induces Streptococcus pneumoniae biofilm dis-persion and transition from asymptomatic colonization to disease. mBio 2013, 4, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Fleming, D.; Rumbaugh, K. The Consequences of Biofilm Dispersal on the Host. Sci. Rep. 2018, 8, 10738. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.P.; Hartmann, A.; Gao, X.; Borriss, R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—A review. Front. Microbiol. 2015, 6, 780. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Wang, C.; Song, X.; Ding, X.; Wu, L.; Wu, H.; Gao, X.; Borriss, R. Bacillus velezensis FZB42 in 2018: The Gram-Positive Model Strain for Plant Growth Promotion and Biocontrol. Front. Microbiol. 2018, 9, 2491. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative analysis of the complete genome sequence of the plant growth–promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef]
- López, D.; Fischbach, M.A.; Chu, F.; Losick, R.; Kolter, R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2009, 106, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Banse, A.V.; Hobbs, E.C.; Losick, R. Phosphorylation of Spo0A by the Histidine Kinase KinD Requires the Lipoprotein Med in Bacillus subtilis. J. Bacteriol. 2011, 193, 3949–3955. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wu, L.; Li, X.; Ma, L.; Borriss, R.; Gao, X. Zn(II) suppresses biofilm formation in Bacillus amyloliquefaciens by inactivation of the Mn(II) uptake. Environ. Microbiol. 2020, 22, 1547–1558. [Google Scholar] [CrossRef] [PubMed]
- Omer Bendori, S.; Pollak, S.; Hizi, D.; Eldar, A. The RapP-PhrP Quorum-Sensing System of Bacillus subtilis Strain NCIB3610 Affects Biofilm Formation through Multiple Targets, Due to an Atypical Signal-Insensitive Allele of RapP. J. Bacteriol. 2015, 197, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qi, J.; Wang, Y.; Wen, J.; Zhao, X.; Qi, G. Comparative study of the role of surfactin-triggered signalling in biofilm formation among different Bacillus species. Microbiol. Res. 2022, 254, 126920. [Google Scholar] [CrossRef]
- Bartolini, M.; Cogliati, S.; Vileta, D.; Bauman, C.; Rateni, L.; Leñini, C.; Argañaraz, F.; Francisco, M.; Villalba, J.M.; Steil, L.; et al. Regulation of Biofilm Aging and Dispersal in Bacillus subtilis by the Alternative Sigma Factor SigB. J. Bacteriol. 2019, 201, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Wang, C.; Ding, X.; Zhu, B.; Song, X.; Borriss, R. AmyloWiki: An integrated database for Bacillus velezensis FZB42, the model strain for plant growth-promoting Bacilli. Database 2019, 2019, baz071. [Google Scholar] [PubMed]
- Barraud, N.; Kjelleberg, S.; Rice, S.A. Dispersal from Microbial Biofilms. Microbiol. Spectr. 2015, 3, 343–362. [Google Scholar] [CrossRef] [PubMed]
- Guilhen, C.; Forestier, C.; Balestrino, D. Biofilm dispersal: Multiple elaborate strategies for dissemination of bacteria with unique properties. Mol. Microbiol. 2017, 105, 188–210. [Google Scholar] [CrossRef]
- Kaplan, J.B. Biofilm Dispersal: Mechanisms, Clinical Implications, and Potential Therapeutic Uses. J. Dent. Res. 2010, 89, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Stacy, A.; Everett, J.; Jorth, P.; Trivedi, U.; Rumbaugh, K.P.; Whiteley, M. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc. Natl. Acad. Sci. USA 2014, 111, 7819–7824. [Google Scholar] [CrossRef]
- Chai, Y.; Norman, T.; Kolter, R.; Losick, R. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev. 2010, 24, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Abee, T.; Kovács, T.; Kuipers, O.P.; van der Veen, S. Biofilm formation and dispersal in Gram-positive bacteria. Curr. Opin. Biotechnol. 2011, 22, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Vlamakis, H.; Aguilar, C.; Losick, R.; Kolter, R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008, 22, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Chua, S.L. Demolishing the great wall of biofilms in Gram-negative bacteria: To disrupt or disperse? Med. Res. Rev. 2020, 40, 1103–1116. [Google Scholar] [CrossRef]
- Ha, D.-G.; O’Toole, G.A. c-di-GMP and its Effects on Biofilm Formation and Dispersion: A Pseudomonas Aeruginosa Review. Microbiol. Spectr. 2015, 3, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Romling, U.; Galperin, M.Y. Discovery of the Second Messenger Cyclic di-GMP. Methods Mol. Biol. 2017, 1657, 1–8. [Google Scholar]
- McDougald, D.; Rice, S.A.; Barraud, N.; Steinberg, P.D.; Kjelleberg, S. Should we stay or should we go: Mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 2012, 10, 39–50. [Google Scholar] [CrossRef]
- Cai, Y.-M.; Hutchin, A.; Craddock, J.; Walsh, M.A.; Webb, J.S.; Tews, I. Differential impact on motility and biofilm dispersal of closely related phosphodiesterases in Pseudomonas aeruginosa. Sci. Rep. 2020, 10, 6232. [Google Scholar] [CrossRef] [PubMed]
- Sprecher, K.S.; Hug, I.; Nesper, J.; Potthoff, E.; Mahi, M.-A.; Sangermani, M.; Kaever, V.; Schwede, T.; Vorholt, J.; Jenal, U. Cohesive Properties of the Caulobacter crescentus Holdfast Adhesin Are Regulated by a Novel c-di-GMP Effector Protein. mBio 2017, 8, e00294-17. [Google Scholar] [CrossRef] [PubMed]
- Nieto, V.; Partridge, J.D.; Severin, G.B.; Lai, R.-Z.; Waters, C.M.; Parkinson, J.S.; Harshey, R.M. Under Elevated c-di-GMP in Escherichia coli, YcgR Alters Flagellar Motor Bias and Speed Sequentially, with Additional Negative Control of the Flagellar Regulon via the Adaptor Protein RssB. J. Bacteriol. 2019, 202, 10–1128. [Google Scholar] [CrossRef]
- Bassis, C.M.; Visick, K.L. The Cyclic-di-GMP Phosphodiesterase BinA Negatively Regulates Cellulose-Containing Biofilms in Vibrio fischeri. J. Bacteriol. 2010, 192, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Su, T.; Wu, H.; Liu, S.; Wang, D.; Zhao, T.; Jin, Z.; Du, W.; Zhu, M.-J.; Chua, S.L.; et al. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 2015, 25, 1352–1367. [Google Scholar] [CrossRef] [PubMed]
- Fagerlund, A.; Smith, V.; Røhr, K.; Lindbäck, T.; Parmer, M.P.; Andersson, K.K.; Reubsaet, L.; Økstad, O.A. Cyclic diguanylate regulation of Bacillus cereus group biofilm formation. Mol. Microbiol. 2016, 101, 471–494. [Google Scholar] [CrossRef]
- Gao, X.; Mukherjee, S.; Matthews, P.M.; Hammad, L.A.; Kearns, D.B.; Dann, C.E. Functional Characterization of Core Components of the Bacillus subtilis Cyclic-Di-GMP Signaling Pathway. J. Bacteriol. 2013, 195, 4782–4792. [Google Scholar] [CrossRef]
- Yang, Y.; Li, Y.; Gao, T.; Zhang, Y.; Wang, Q. C-di-GMP turnover influences motility and biofilm formation in Bacillus amyloliquefaciens PG12. Res. Microbiol. 2018, 169, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Yu, Z.; Liu, S.; Chen, B.; Zhu, L.; Li, Z.; Chou, S.-H.; He, J. c-di-GMP Regulates Various Phenotypes and Insecticidal Activity of Gram-Positive Bacillus thuringiensis. Front. Microbiol. 2018, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [PubMed]
- Rumbaugh, K.P.; Sauer, K. Biofilm dispersion. Nat. Rev. Microbiol. 2020, 18, 571–586. [Google Scholar] [CrossRef]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef]
- Yaryura, P.M.; León, M.; Correa, O.S.; Kerber, N.L.; Pucheu, N.L.; García, A.F. Assessment of the Role of Chemotaxis and Biofilm Formation as Requirements for Colonization of Roots and Seeds of Soybean Plants by Bacillus amyloliquefaciens BNM339. Curr. Microbiol. 2008, 56, 625–632. [Google Scholar] [CrossRef]
- Fan, B.; Borriss, R.; Bleiss, W.; Wu, X. Gram-positive rhizobacterium Bacillus amyloliquefaciens FZB42 colonizes three types of plants in different patterns. J. Microbiol. 2012, 50, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Al-Ali, A.; Deravel, J.; Krier, F.; Béchet, M.; Ongena, M.; Jacques, P. Biofilm formation is determinant in tomato rhizosphere colonization by Bacillus velezensis FZB42. Environ. Sci. Pollut. Res. 2018, 25, 29910–29920. [Google Scholar] [CrossRef]
- Weng, J.; Wang, Y.; Li, J.; Shen, Q.; Zhang, R. Enhanced root colonization and biocontrol activity of Bacillus amyloliquefaciens SQR9 by abrB gene disruption. Appl. Microbiol. Biotechnol. 2013, 97, 8823–8830. [Google Scholar] [CrossRef]
- Auger, S.; Krin, E.; Aymerich, S.; Gohar, M. Autoinducer 2 Affects Biofilm Formation by Bacillus cereus. Appl. Environ. Microbiol. 2006, 72, 937–941. [Google Scholar] [CrossRef]
- Kolodkin-Gal, I.; Romero, D.; Cao, S.; Clardy, J.; Kolter, R.; Losick, R. d-Amino Acids Trigger Biofilm Disassembly. Science 2010, 328, 627–629. [Google Scholar] [CrossRef] [PubMed]
- Schiött, T.; von Wachenfeldt, C.; Hederstedt, L. Identification and characterization of the ccdA gene, required for cytochrome c synthesis in Bacillus subtilis. J. Bacteriol. 1997, 179, 1962–1973. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, M.; Kobayashi, K. Calcium Prevents Biofilm Dispersion in Bacillus subtilis. J. Bacteriol. 2021, 203, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Reuter, M.; Ultee, E.; Toseafa, Y.; Tan, A.; van Vliet, A.H.M. Inactivation of the core cheVAWY chemotaxis genes disrupts chemotactic motility and organised biofilm formation in Campylobacter jejuni. FEMS Microbiol. Lett. 2020, 367, fnaa198. [Google Scholar] [CrossRef] [PubMed]
- Conrad, J.C. Physics of bacterial near-surface motility using flagella and type IV pili: Implications for biofilm formation. Res. Microbiol. 2012, 163, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Gu, Y.; Chen, G.; Wang, G.; Liu, L. Flagellar motility mediates early-stage biofilm formation in oligotrophic aquatic environment. Ecotoxicol. Environ. Saf. 2020, 194, 110340. [Google Scholar] [CrossRef] [PubMed]
- Pawar, S.V.; Messina, M.; Rinaldo, S.; Cutruzzolà, F.; Kaever, V.; Rampioni, G.; Leoni, L. Novel genetic tools to tackle c-di-GMP-dependent signalling in Pseudomonas aeruginosa. J. Appl. Microbiol. 2016, 120, 205–217. [Google Scholar] [CrossRef]
- Lin Chua, S.; Liu, Y.; Li, Y.; Jun Ting, H.; Kohli, G.S.; Cai, Z.; Suwanchaikasem, P.; Kau Kit Goh, K.; Pin Ng, S.; Tolker-Nielsen, T.; et al. Reduced Intracellular c-di-GMP Content Increases Expression of Quorum Sensing-Regulated Genes in Pseu-domonas aeruginosa. Front. Cell. Infect. Microbiol. 2017, 7, 451. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.B.; Kragh, K.N.; Hultqvist, L.D.; Rybtke, M.; Nilsson, M.; Jakobsen, T.H.; Givskov, M.; Tolker-Nielsen, T. Induction of Native c-di-GMP Phosphodiesterases Leads to Dispersal of Pseudomonas aeruginosa Biofilms. Antimicrob. Agents Chemother. 2021, 65, 10–1128. [Google Scholar] [CrossRef]
- Hull, T.D.; Ryu, M.-H.; Sullivan, M.J.; Johnson, R.C.; Klena, N.T.; Geiger, R.M.; Gomelsky, M.; Bennett, J.A. Cyclic Di-GMP Phosphodiesterases RmdA and RmdB Are Involved in Regulating Colony Morphology and Development in Streptomyces coelicolor. J. Bacteriol. 2012, 194, 4642–4651. [Google Scholar] [CrossRef]
- Gao, X. Exploration of C-di-GMP Signaling Pathway in B. subtilis. Ph.D. Thesis, Indiana University, Bloomington, IN, USA, 2014. [Google Scholar]
- Le Breton, Y.; Mohapatra, N.P.; Haldenwang, W.G. In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Appl. Environ. Microbiol. 2006, 72, 327–333. [Google Scholar] [CrossRef]
- Dietel, K.; Beator, B.; Budiharjo, A.; Fan, B.; Borriss, R. Bacterial Traits Involved in Colonization of Arabidopsis thaliana Roots by Bacillus amyloliquefaciens FZB42. Plant Pathol. J. 2013, 29, 59–66. [Google Scholar] [CrossRef]
- Möller, E.; Bahnweg, G.; Sandermann, H.; Geiger, H. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 1992, 20, 6115–6116. [Google Scholar] [CrossRef]
- Fan, B.; Chen, X.H.; Budiharjo, A.; Bleiss, W.; Vater, J.; Borriss, R. Efficient colonization of plant roots by the plant growth promoting bacterium Bacillus amyloliquefaciens FZB42, engineered to express green fluorescent protein. J. Biotechnol. 2011, 151, 303–311. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




| Strain | Genotype | Description | Reference/Source |
|---|---|---|---|
| Plasmids | |||
| pMarA | pUC19 carrying TnYLB-1 transposon mariner Himar1 transposase and promoters σA, KmR ApR EmR | Le Breton et al., 2006 [62] | |
| pMD-19 | Commercial T-Vector (AmpR) | Takara, Maebashi, Japan | |
| pFB01 | pVBF-amyE::emR, gfp+ | Lab stock | |
| pFB103 | pMD-19-speR | Lab stock | |
| pSZZ11 | pMD-19-ccdC | This work | |
| pSZZ13 | pFB01-ccdC | This work | |
| pSZZ17 | pMD-19-ccdC-speR | This work | |
| FZB42 | Wild-type | Lab stock | |
| SZZ06 | FZB42 ccdC::TnYLB-1 | Impaired in biofilm dispersal | This work |
| FBS239 | pMarA → FZB42 | Laboratory preservation | |
| SZZ15 | SZZ06 amyE::ccdC, Emr | This work | |
| SZZ18 | FZB42 ccdC::speR | pSZZ17 → FZB42 | This work |
| Primer | Sequence (5′ to 3′) |
|---|---|
| FBO752 | GCTTGTAAATTCTATCATAATTG |
| FBO753 | AGGGAATCATTTGAAGGTTGG |
| szz01 | AAGCATCTAAAGTGCTGGAG |
| szz02 | CAGATTGATCTTACTCCTTATC |
| szz38 | ATCGGTCTTGCGTTTGCAG |
| szz39 | TGTCCGGTCATATCAGTCAT |
| szz44 + Kpn I | CGGGGTACCATGATGATTATAATTTCATCCG |
| szz45 + Cla I | CCCATCGATATTCATCTGAATATCAGCGG |
| szz46 + Nru I | GTCTCGCGAGCATATGATCAGATCTTAAGGCC |
| szz47 + Nru I | GTCTCGCGATTGAAGCATGCAAATGTCACT |
| FBO16 | TGGGTCAATCGAGAATATCGTC |
| FBO550 | GTTTGTCTGCCGTGATGT |
| gyrA-fr | CCCACGTCCTCATAGTGACAG |
| gyrA-re | CGGACCGTTGCTGTCAGTGA |
| ydaK-fr | TGAAACAGCTGCGTGAAGAG |
| ydaK-re | AAGGAATTGCCGTAGCGTTC |
| yhcK-fr | AATCAGGCACACGATTGACG |
| yhcK-re | TTCGCTTTGTACAGCATCCG |
| ykuI-fr | CGCTGAGGAACAAAGAGTCG |
| yukI-re | CAATCTCCGCATCTGCCAAA |
| ytrP-fr | CGGAGCATAATGCGGTTCAT |
| ytrP-re | AGTGACGGCCTTTCATCAGA |
| yuxH-fr | TTCTTCACAGGGAGCTGGAG |
| yuxH-re | CGTCACAGTCCCAGTTGTTG |
| yybT-fr | GATAACTGGGTTGCCGCATT |
| yybT-re | TTTGACCAGGTGCTGATTGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, L.; Shen, Z.; Li, M.; Guan, C.; Fan, B.; Chai, Y.; Zhao, Y. ccdC Regulates Biofilm Dispersal in Bacillus velezensis FZB42. Int. J. Mol. Sci. 2024, 25, 5201. https://doi.org/10.3390/ijms25105201
Shao L, Shen Z, Li M, Guan C, Fan B, Chai Y, Zhao Y. ccdC Regulates Biofilm Dispersal in Bacillus velezensis FZB42. International Journal of Molecular Sciences. 2024; 25(10):5201. https://doi.org/10.3390/ijms25105201
Chicago/Turabian StyleShao, Lin, Zizhu Shen, Meiju Li, Chenyun Guan, Ben Fan, Yunrong Chai, and Yinjuan Zhao. 2024. "ccdC Regulates Biofilm Dispersal in Bacillus velezensis FZB42" International Journal of Molecular Sciences 25, no. 10: 5201. https://doi.org/10.3390/ijms25105201





