Peripheral Blood Gene Expression Profiling Reveals Molecular Pathways Associated with Cervical Artery Dissection
Abstract
:1. Introduction
- 1.
- Disorganized thinning and straightening of the internal elastic membrane.
- 2.
- Heterogeneous thickening of the tunica media with areas of non-inflammatory mucoid degeneration, necrosis, fibrosis, and calcification.
- 3.
- Predominance of smooth-muscle cells with a synthetic phenotype (vacuolar degeneration, fissed mitochondria, and irregular cell polarization).
- 4.
- Erythrocyte extravasation (due to rupture of the vasa vasorum) and neovascularization foci on the media-adventitia border.
- 5.
- Electron microscopy signs of mitochondrial cytopathy (circular cristae and pathological inclusions in the matrix of mitochondria) and changes in the structure of extracellular elastic and collagen fibers.
2. Results
2.1. Differential Gene Expression Analysis with DAVID Functional Annotation
2.2. Gene–Disease Associations
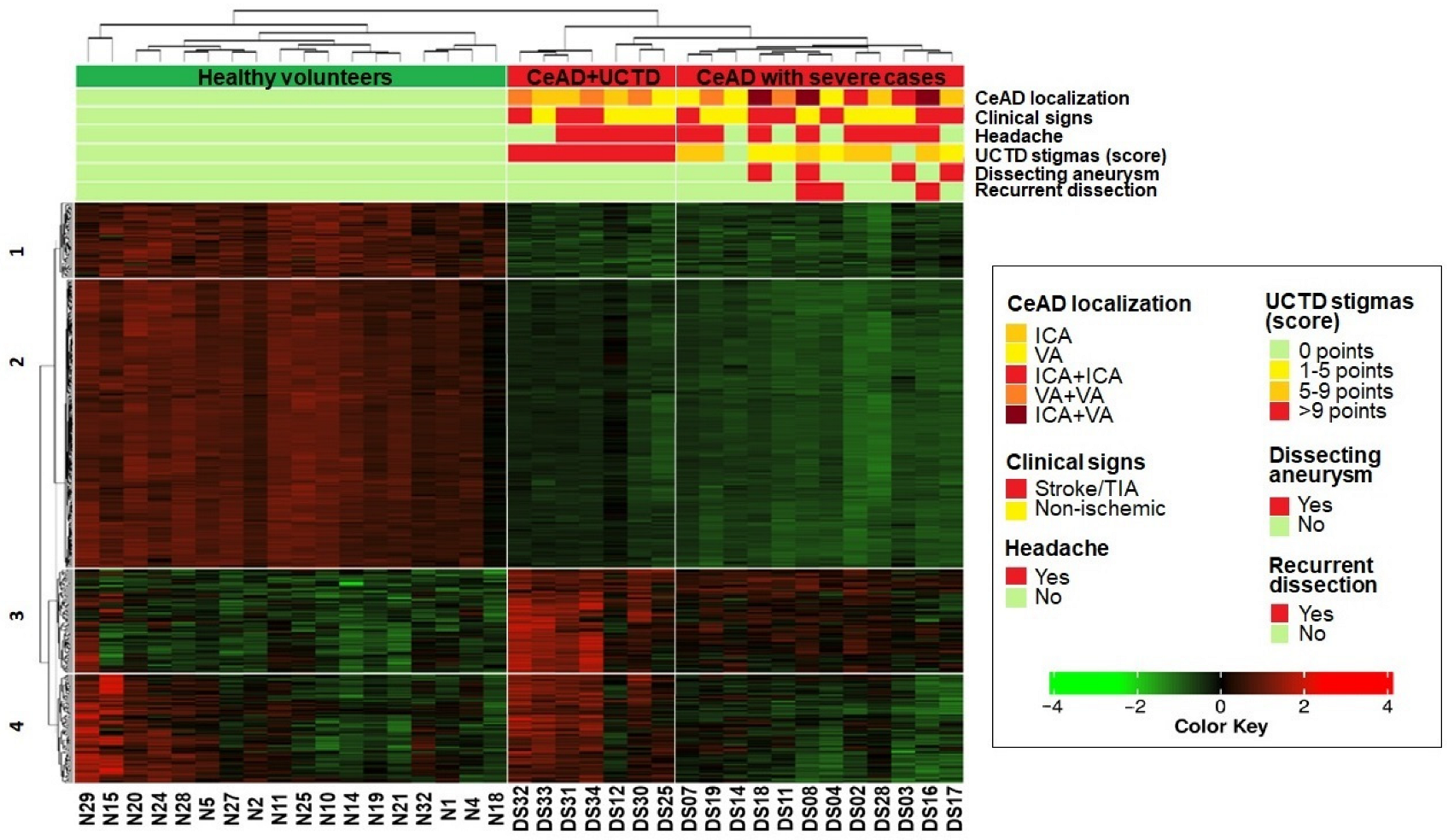
2.3. Heatmap Clustering
- 1.
- Healthy volunteers,
- 2.
- Patients with the most pronounced stigmas of UCTD (>9 points),
- 3.
- Patients, including all the most severe cases of CeAD (recurrent dissection, ICA/VA aneurysm, and combined ICA/VA dissection).
- The first and second clusters (63 and 246 genes, respectively) contained genes with downregulated expression in both CeAD subgroups that were enriched in cell cycle regulation, translation, mitochondrial respiration, and ribosome processing (FDR < 0.05).
- The third cluster comprised 87 genes that were upregulated in both CeAD subgroups and enriched in calcium metabolism, G-protein-associated signaling, and the regulation of cell polarization and migration (FDR < 0.05).
- In the fourth cluster, 91 genes were only upregulated in the subgroup of patients with the most pronounced stigmas of UCTD and in some healthy volunteers. These genes were related to phagocytosis, the exchange of membrane lipids, and VEGFA-VEGFR2 signaling (FDR < 0.05).
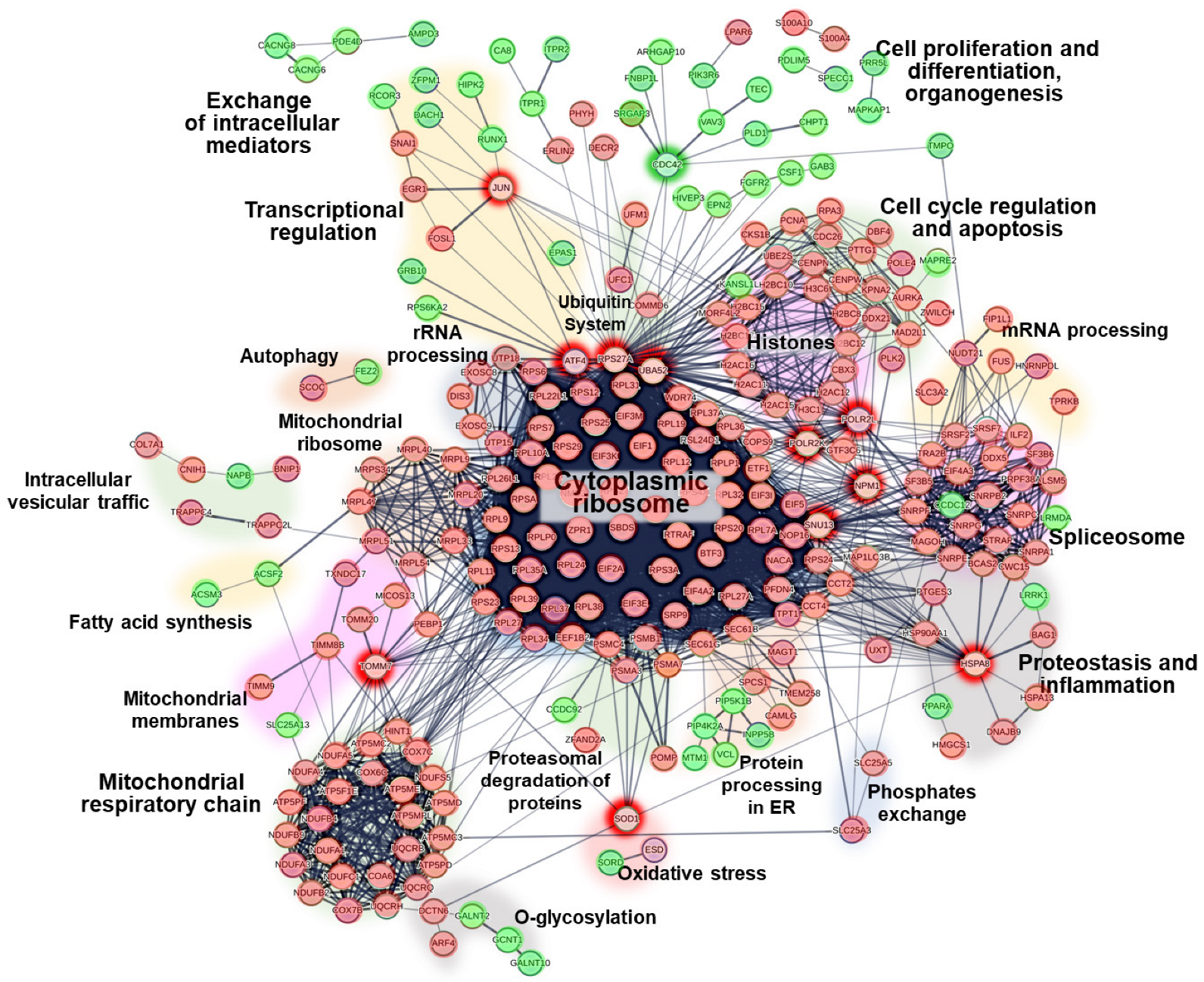
2.4. Protein–Protein Interactions
2.5. Gene Set Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. Clinical Description of the Cohort
- 1.
- A dissection of two or more cervical arteries (10 cases).
- 2.
- A recurrent dissection during 1 year of observation after the primary CeAD (3 cases).
- 3.
- An intense chronic headache presenting long before the development of CeAD (13 cases).
- 4.
- A dissecting aneurysm of the VA and/or ICA diagnosed with MR angiography (4 cases).
- 5.
4.2. Sample Processing
4.3. Bioinformatics Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dobrynina, L.A.; Kalashnikova, L.A.; Pavlova, L.N. Ischemic stroke in young age. Zhurnal Nevrol. Psikhiatrii Im. S.S. Korsakova 2011, 111, 4–8. (In Russian) [Google Scholar] [CrossRef]
- Blum, C.A.; Yaghi, S. Cervical Artery Dissection: A Review of the Epidemiology, Pathophysiology, Treatment, and Outcome. Arch. Neurosci. 2015, 2, e26670. [Google Scholar] [CrossRef] [PubMed]
- Debette, S.; Leys, D. Cervical-artery dissections, predisposing factors, diagnosis, and outcome. Lancet Neurol. 2009, 8, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Keser, Z.; Meschia, J.F.; Lanzino, G. Craniocervical Artery Dissections, A Concise Review for Clinicians. Mayo Clin. Proc. 2022, 97, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Kalashnikova, L.A.; Dobrynina, L.A. Dissection of internal carotid and vertebral arteries, clinical presentation, diagnosis, and treatment. Neurol. Neuropsychiatry Psychosom. 2013, 5 (Suppl. S2), 40–45. (In Russian) [Google Scholar] [CrossRef]
- Kalashnikova, L.; Gulevskaya, T.; Sakharova, A.; Chaykovskaya, R.; Gubanova, M.; Danilova; Shabalina, A.; Dobrynina, L. Internal carotid and vertebral artery dissection, morphology, pathophysiology and provoking factors. Bull. RSMU 2019, 5, 78–85. (In Russian) [Google Scholar] [CrossRef]
- Anadure, R.K.; Mohimen, A.; Saxena, R.; Sivasankar, R. A Study on the Clinical and Angiographic Spectrum of Spontaneous Extracranial Dissections in the Cerebral Vasculature. J. Neurosci. Rural Pract. 2018, 9, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Markus, H.S.; Levi, C.; King, A.; Madigan, J.; Norris, J. Cervical Artery Dissection in Stroke Study (CADISS) Investigators. Antiplatelet Therapy vs. Anticoagulation Therapy in Cervical Artery Dissection, The Cervical Artery Dissection in Stroke Study (CADISS) Randomized Clinical Trial Final Results. JAMA Neurol. 2019, 76, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Keser, Z.; Chiang, C.C.; Benson, J.C.; Pezzini, A.; Lanzino, G. Cervical Artery Dissections, Etiopathogenesis and Management. Vasc. Health Risk Manag. 2022, 18, 685–700. [Google Scholar] [CrossRef]
- Dittrich, R.; Heidbreder, A.; Rohsbach, D.; Schmalhorst, J.; Nassenstein, I.; Maintz, D.; Ringelstein, E.B.; Nabavi, D.G.; Kuhlenbäumer, G. Connective tissue and vascular phenotype in patients with cervical artery dissection. Neurology 2007, 68, 2120–2124. [Google Scholar] [CrossRef]
- Del Zotto, E.; Grassi, M.; Zedde, M.; Zini, A.; Bersano, A.; Gandolfo, C.; Silvestrelli, G.; Baracchini, C.; Cerrato, P.; Lodigiani, C.; et al. Risk Profile of Patients with Spontaneous Cervical Artery Dissection. Ann. Neurol. 2023, 94, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Brandt, T.; Hausser, I.; Orberk, E.; Grau, A.; Hartschuh, W.; Anton-Lamprecht, I.; Hacke, W. Ultrastructural connective tissue abnormalities in patients with spontaneous cervicocerebral artery dissections. Ann. Neurol. 1998, 44, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Brandt, T.; Orberk, E.; Weber, R.; Werner, I.; Busse, O.; Müller, B.T.; Wigger, F.; Grau, A.; Grond–Ginsbach, C.; Hausser, I. Pathogenesis of cervical artery dissections, association with connective tissue abnormalities. Neurology 2001, 57, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Kalashnikova, L.A.; Gulevskaya, T.S.; Anufriev, P.L.; Gnedovskaya, E.V.; Konovalov, R.N.; Piradov, M.A. Ischemic stroke in young age due to dissection of intracranial carotid artery and its branches (clinical and morphological study). Ann. Clin. Exp. Neurol. 2009, 3, 18–24. (In Russian) [Google Scholar] [CrossRef]
- Kalashnikova, L.A.; Chaykovskaya, R.P.; Dobrynina, L.A.; Sakharova, A.V.; Gulevskaya, T.S.; Dreval, M.V.; Ivanova, M.V. Internal carotid artery dissection as a cause of severe ischemic stroke with lethal outcome. Zhurnal Nevrol. I Psikhiatrii Im. S.S. Korsakova 2015, 115, 19–25. (In Russian) [Google Scholar] [CrossRef]
- Bax, M.; Romanov, V.; Junday, K.; Giannoulatou, E.; Martinac, B.; Kovacic, J.C.; Liu, R.; Iismaa, S.E.; Graham, R.M. Arterial dissections, Common features and new perspectives. Front. Cardiovasc. Med. 2022, 9, 1055862. [Google Scholar] [CrossRef] [PubMed]
- Lesauskaite, V.; Tanganelli, P.; Sassi, C.; Neri, E.; Diciolla, F.; Ivanoviene, L.; Epistolato, M.C.; Lalinga, A.V.; Alessandrini, C.; Spina, D. Smooth muscle cells of the media in the dilatative pathology of ascending thoracic aorta, morphology, immunoreactivity for osteopontin, matrix metalloproteinases, and their inhibitors. Hum. Pathol. 2001, 32, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, J.; Fu, W.; Guo, D.; Jiang, J.; Wang, Y. Association of smooth muscle cell phenotypes with extracellular matrix disorders in thoracic aortic dissection. J. Vasc. Surg. 2012, 56, 1698–1709. [Google Scholar] [CrossRef]
- Hao, H.; Gabbiani, G.; Bochaton-Piallat, M.L. Arterial smooth muscle cell heterogeneity, implications for atherosclerosis and restenosis development. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 1510–1520. [Google Scholar] [CrossRef]
- Debette, S.; Kamatani, Y.; Metso, T.M.; Kloss, M.; Chauhan, G.; Engelter, S.T.; Pezzini, A.; Thijs, V.; Markus, H.S.; Dichgans, M.; et al. Common variation in PHACTR1 is associated with susceptibility to cervical artery dissection. Nat. Genet. 2015, 47, 78–83. [Google Scholar] [CrossRef]
- Daghlas, I.; Sargurupremraj, M.; Danning, R.; Gormley, P.; Malik, R.; Amouyel, P.; Metso, T.; Pezzini, A.; Kurth, T.; Debette, S.; et al. Migraine, Stroke, and Cervical Arterial Dissection, Shared Genetics for a Triad of Brain Disorders with Vascular Involvement. Neurol. Genet. 2022, 8, e653. [Google Scholar] [CrossRef]
- Debette, S.; Markus, H.S. The genetics of cervical artery dissection, a systematic review. Stroke 2009, 40, 459–466. [Google Scholar] [CrossRef]
- Techlo, T.R.; Rasmussen, A.H.; Møller, P.L.; Bøttcher, M.; Winther, S.; Davidsson, O.B.; Olofsson, I.A.; Chalmer, M.A.; Kogelman, L.J.A.; Nyegaard, M.; et al. Familial analysis reveals rare risk variants for migraine in regulatory regions. Neurogenetics 2020, 21, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Sopić, M.; Karaduzovic-Hadziabdic, K.; Kardassis, D.; Maegdefessel, L.; Martelli, F.; Meerson, A.; Munjas, J.; Niculescu, L.S.; Stoll, M.; Magni, P.; et al. Transcriptomic research in atherosclerosis, Unravelling plaque phenotype and overcoming methodological challenges. J. Mol. Cell. Cardiol. Plus 2023, 6, 100048. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, X.; Huo, B.; Feng, X.; Fang, Z.-M.; Jiang, D.-S.; Wei, X. Integrating Bulk Transcriptome and Single-Cell RNA Sequencing Data Reveals the Landscape of the Immune Microenvironment in Thoracic Aortic Aneurysms. Front. Cardiovasc. Med. 2022, 9, 846421. [Google Scholar] [CrossRef]
- Chou, E.L.; Chaffin, M.; Simonson, B.; Pirruccello, J.P.; Akkad, A.-D.; Nekoui, M.; Cardenas, C.L.L.; Bedi, K.C.; Nash, C.; Juric, D.; et al. Aortic Cellular Diversity and Quantitative Genome-Wide Association Study Trait Prioritization Through Single-Nuclear RNA Sequencing of the Aneurysmal Human Aorta. Arterioscler. Thromb. Vasc. Biol. 2022, 42, 1355–1374. [Google Scholar] [CrossRef]
- Hautakangas, H.; Winsvold, B.S.; Ruotsalainen, S.E. International Headache Genetics Consortium, HUNT All-in Headache, Danish Blood Donor Study Genomic Cohort; et al. Genome-wide analysis of 102,084 migraine cases identifies 123 risk loci and subtype-specific risk alleles. Nat. Genet. 2022, 54, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Brajanovski, N.; Chan, K.T.; Xuan, J.; Pearson, R.B.; Sanij, E. Ribosomal proteins and human diseases, molecular mechanisms and targeted therapy. Signal. Transduct. Target. Ther. 2021, 6, 323. [Google Scholar] [CrossRef] [PubMed]
- Farley-Barnes, K.I.; Ogawa, L.M.; Baserga, S.J. Ribosomopathies, Old Concepts, New Controversies. Trends Genet. 2019, 35, 754–767. [Google Scholar] [CrossRef]
- Warner, J.R.; McIntosh, K.B. How common are extraribosomal functions of ribosomal proteins? Mol. Cell. 2009, 34, 3–11. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, W.J.; Liao, J.M.; Liao, P.; Lu, H. Ribosomal proteins, functions beyond the ribosome. J. Mol. Cell Biol. 2015, 7, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Yang, J.; Yi, J. Nucleolar Stress, hallmarks, sensing mechanism and diseases. Cell Stress 2018, 2, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Lafita-Navarro, M.C.; Conacci-Sorrell, M. Nucleolar stress, From development to cancer. Semin. Cell Dev. Biol. 2023, 136, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Bastide, A.; David, A. The ribosome, (slow) beating heart of cancer (stem) cell. Oncogenesis 2018, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, J. Ribosome heterogeneity in stem cells and development. J Cell Biol. 2020, 219, e202001108. [Google Scholar] [CrossRef] [PubMed]
- Gonskikh, Y.; Polacek, N. Alterations of the translation apparatus during aging and stress response. Mech. Ageing Dev. 2017, 168, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Teng, T.; Thomas, G.; Mercer, C.A. Growth control and ribosomopathies. Curr. Opin. Genet. Dev. 2013, 23, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Maehama, T.; Nishio, M.; Otani, J.; Mak, T.W.; Suzuki, A. Nucleolar stress, Molecular mechanisms and related human diseases. Cancer Sci. 2023, 114, 2078–2086. [Google Scholar] [CrossRef] [PubMed]
- Pfister, A.S. Emerging Role of the Nucleolar Stress Response in Autophagy. Front. Cell Neurosci. 2019, 13, 156. [Google Scholar] [CrossRef]
- Zhang, W.; Cheng, W.; Parlato, R.; Guo, X.; Cui, X.; Dai, C.; Xu, L.; Zhu, J.; Zhu, M.; Luo, K.; et al. Nucleolar stress induces a senescence-like phenotype in smooth muscle cells and promotes development of vascular degeneration. Aging 2020, 12, 22174–22198. [Google Scholar] [CrossRef]
- Aladesuyi Arogundade, O.; Nguyen, S.; Leung, R.; Wainio, D.; Rodriguez, M.; Ravits, J. Nucleolar stress in C9orf72 and sporadic ALS spinal motor neurons precedes TDP-43 mislocalization. Acta Neuropathol. Commun. 2021, 9, 26. [Google Scholar] [CrossRef]
- Kang, H.; Shin, J.H. Repression of rRNA transcription by PARIS contributes to Parkinson’s disease. Neurobiol. Dis. 2015, 73, 220–228. [Google Scholar] [CrossRef]
- Nyhus, C.; Pihl, M.; Hyttel, P.; Hall, V.J. Evidence for nucleolar dysfunction in Alzheimer’s disease. Rev. Neurosci. 2019, 30, 685–700. [Google Scholar] [CrossRef]
- Sönmez, A.; Mustafa, R.; Ryll, S.T.; Tuorto, F.; Wacheul, L.; Ponti, D.; Litke, C.; Hering, T.; Kojer, K.; Koch, J.; et al. Nucleolar stress controls mutant Huntington toxicity and monitors Huntington’s disease progression. Cell Death Dis. 2021, 12, 1139. [Google Scholar] [CrossRef]
- Tsoi, H.; Lau, T.C.K.; Tsang, S.Y.; Lau, K.F.; Chan, H.Y.E. CAG expansion induces nucleolar stress in polyglutamine diseases. Proc. Natl. Acad. Sci. USA 2012, 109, 13428–13433. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, A.; Pagano, M.; Russo, G.; Russo, A. Ribosome Biogenesis and Cancer, Overview on Ribosomal Proteins. Int J Mol Sci. 2021, 22, 5496. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, A.; Hasan, S.A.; Chawla, L.; Barzallo, P.; Bhardwaj, C. Spontaneous coronary artery dissection in a patient on platinum-based chemotherapy for testicular malignancy. J. Am. Coll. Cardiol. 2023, 81 (Suppl. S8), 2712. [Google Scholar] [CrossRef]
- Somov, P.; Marchak, D.; Matusov, A.; Viller, A.; Shevchenko, Y.; Miminoshvili, A. Spontaneous coronary artery dissection during cisplatin and capecitabine therapy. Ann. Med. Surg. 2019, 45, 1–5. [Google Scholar] [CrossRef]
- Van Chien, D.; Hai, P.D.; Nhung, L.T.; Son, P.T. Multiple spontaneous coronary artery dissections associated with intravenous daunorubicin treatment for acute myelocytic leukaemia, a case report. Eur. Heart J. Case Rep. 2021, 5, 427. [Google Scholar] [CrossRef]
- Khurana, S.; Kaushal, J.; Markan, D.; Wang, J.C.; Kaliyadan, A.; Haas, C.; Devkota, A. Recurrent Spontaneous Coronary Artery Dissection, A Case of Triple Trouble. J. Community Hosp. Intern. Med. Perspect. 2023, 13, 63–67. [Google Scholar] [CrossRef]
- Mantia-Smaldone, G.M.; Bagley, L.J.; Kasner, S.E.; Chu, S. Vertebral artery dissection and cerebral infarction in a patient with recurrent ovarian cancer receiving bevacizumab. Gynecol. Oncol. Case Rep. 2013, 5, 37–39. [Google Scholar] [CrossRef] [PubMed]
- Groden, P.J.; Lee, T.C.; Bhattacharyya, S.; Connors, J.; Lorch, J. Lenvatinib-Associated Cervical Artery Dissections in a Patient with Radioiodine-Refractory Metastatic Papillary Thyroid Carcinoma. Front. Med. 2017, 4, 220. [Google Scholar] [CrossRef] [PubMed]
- Khadjooi, K.; Adab, N.; Kenton, A. Acute stroke secondary to carotid artery dissection in a patient with germ cell tumour, did cisplatin play a role? Onkologie 2013, 36, 46–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, M.; Zhang, X.; Zhang, L.; Jia, M.; Shen, Z.; Wang, J.; Zhao, B.; Gong, Y.; Gong, J. Aneurysm and Artery Dissection Following the Use of Vascular Endothelial Growth Factor Inhibitor, A Real-World Analysis Using a Spontaneous Reporting System. J. Am. Heart Assoc. 2021, 10, e020844. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Yeon, B.; Kim, M.S.; Yoo, M.; Kim, B.; Yu, Y.M. Aneurysm and Artery Dissection After Oral VEGFR-TKI Use in Adults With Cancer. JAMA Netw. Open 2023, 6, e2345977. [Google Scholar] [CrossRef] [PubMed]
- Coppé, J.P.; Desprez, P.Y.; Krtolica, A.; Campisi, J. The senescence-associated secretory phenotype, the dark side of tumor suppression. Annu. Rev. Pathol. 2010, 5, 99–118. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Song, T.; Chen, L.; Soliman, H.; Chen, J. Nucleolar repression facilitates initiation and maintenance of senescence. Cell Cycle 2015, 14, 3613–3623. [Google Scholar] [CrossRef] [PubMed]
- Laberge, R.M.; Awad, P.; Campisi, J.; Desprez, P.Y. Epithelial-mesenchymal transition induced by senescent fibroblasts. Cancer Microenviron. 2012, 5, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Faheem, M.M.; Seligson, N.D.; Ahmad, S.M.; Rasool, R.U.; Gandhi, S.G.; Bhagat, M.; Goswami, A. Convergence of therapy-induced senescence (TIS) and EMT in multistep carcinogenesis, current opinions and emerging perspectives. Cell Death Discov. 2020, 6, 51. [Google Scholar] [CrossRef]
- Haerinck, J.; Goossens, S.; Berx, G. The epithelial-mesenchymal plasticity landscape, principles of design and mechanisms of regulation. Nat. Rev. Genet. 2023, 24, 590–609. [Google Scholar] [CrossRef]
- Kovacic, J.C.; Mercader, N.; Torres, M.; Boehm, M.; Fuster, V. Epithelial-to-mesenchymal and endothelial-to-mesenchymal transition, from cardiovascular development to disease. Circulation 2012, 125, 1795–1808. [Google Scholar] [CrossRef] [PubMed]
- Alvandi, Z.; Bischoff, J. Endothelial-Mesenchymal Transition in Cardiovascular Disease. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 2357–2369. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Park, J.Y.; Lee, H.; Song, E.S.; Kuk, M.U.; Joo, J.; Oh, S.; Kwon, H.W.; Park, J.T.; Park, S.C. Targeting Mitochondrial Metabolism as a Strategy to Treat Senescence. Cells 2021, 10, 3003. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Yu, Y.; Kim, J.-H.; Lee, J.; Park, J.; Hong, K.; Seo, J.-K.; Lim, C.; Min, K.-T. Suboptimal Mitochondrial Activity Facilitates Nuclear Heat Shock Responses for Proteostasis and Genome Stability. Mol. Cells 2023, 46, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.N.; Tortelote, G.G.; Pascale, C.L.; McCormack, I.G.; Nordham, K.D.; Suder, N.J.; Couldwell, M.W.; Dumont, A.S. Single-Cell Transcriptome Analysis of the Circle of Willis in a Mouse Cerebral Aneurysm Model. Stroke 2022, 53, 2647–2657. [Google Scholar] [CrossRef] [PubMed]
- Pfister, A.S. An Update on Nucleolar Stress, The Transcriptional Control of Autophagy. Cells 2023, 12, 2071. [Google Scholar] [CrossRef] [PubMed]
- Gubanova, M.V.; Kalashnikova, L.A.; Dobrynina, L.A.; Shamtieva, K.V.; Berdalin, A.B. Markers of connective tissue dysplasia in cervical artery dissection and its predisposing factors. Ann. Clin. Exp. Neurol. 2017, 11, 19–28. (In Russian) [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR, ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR, a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes., revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP, an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID, a web server for functional enrichment analysis and functional annotation of gene lists. Nucleic Acids Res. 2022, 50, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Croft, D.; O’Kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B.; et al. Reactome, a database of reactions., pathways and biological processes. Nucleic Acids Res. 2011, 39 (Suppl. S1), 691–697. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG, kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Mélius, J.; Cirillo, E.; Coort, S.L.; Digles, D.; et al. WikiPathways, a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018, 46, 661–667. [Google Scholar] [CrossRef]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET, a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017, 45, 833–839. [Google Scholar] [CrossRef]
- Gel, B.; Diez-Villanueva, A.; Serra, E.; Buschbeck, M.; Peinado, M.A.; Malinverni, R. regioneR, an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 2016, 32, 289–291. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler, an R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023, protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, 638–646. [Google Scholar] [CrossRef] [PubMed]




| Characteristics | Number of Patients | Ischemic Stroke/Transient Ischemic Attack Due to Dissection | Isolated Headache/Neck Pain Due to Dissection | UCTD Stigmas | Chronic Headache in Anamnesis | ||||
|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | Mean Points Score | N | % | |
| All | 19 | 100 | 9 | 47 | 10 | 53 | 8.9 ± 3.8 | 13 | 68 |
| Women | 13 | 68 | 4 | 21 | 9 | 48 | 10.9 ± 3.7 | 10 | 52 |
| Men | 6 | 32 | 5 | 26 | 1 | 5 | 4.2 ± 3.5 | 3 | 16 |
| ICA dissection | 5 | 26 | 2 | 22 | 3 | 30 | 9.6 ± 3.7 | 3 | 23 |
| VA dissection | 4 | 21 | 2 | 22 | 2 | 20 | 8.6 ± 4.7 | 2 | 15 |
| Dissection of two or more cervical arteries | 10 | 53 | 5 | 32 | 5 | 50 | 8.6 ± 3.5 | 8 | 62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shlapakova, P.S.; Dobrynina, L.A.; Kalashnikova, L.A.; Gubanova, M.V.; Danilova, M.S.; Gnedovskaya, E.V.; Grigorenko, A.P.; Gusev, F.E.; Manakhov, A.D.; Rogaev, E.I. Peripheral Blood Gene Expression Profiling Reveals Molecular Pathways Associated with Cervical Artery Dissection. Int. J. Mol. Sci. 2024, 25, 5205. https://doi.org/10.3390/ijms25105205
Shlapakova PS, Dobrynina LA, Kalashnikova LA, Gubanova MV, Danilova MS, Gnedovskaya EV, Grigorenko AP, Gusev FE, Manakhov AD, Rogaev EI. Peripheral Blood Gene Expression Profiling Reveals Molecular Pathways Associated with Cervical Artery Dissection. International Journal of Molecular Sciences. 2024; 25(10):5205. https://doi.org/10.3390/ijms25105205
Chicago/Turabian StyleShlapakova, Polina S., Larisa A. Dobrynina, Ludmila A. Kalashnikova, Mariia V. Gubanova, Maria S. Danilova, Elena V. Gnedovskaya, Anastasia P. Grigorenko, Fedor E. Gusev, Andrey D. Manakhov, and Evgeny I. Rogaev. 2024. "Peripheral Blood Gene Expression Profiling Reveals Molecular Pathways Associated with Cervical Artery Dissection" International Journal of Molecular Sciences 25, no. 10: 5205. https://doi.org/10.3390/ijms25105205







