Pharmacological Evaluation of Cannabinoid Receptor Modulators Using GRABeCB2.0 Sensor
Abstract
:1. Introduction
2. Results and Discussion
2.1. GRABeCB2.0 Assays for CB1R Agonists
2.2. GRABeCB2.0 Assays for Antagonist/Inverse Agonists
2.3. 3-Arm Modulators
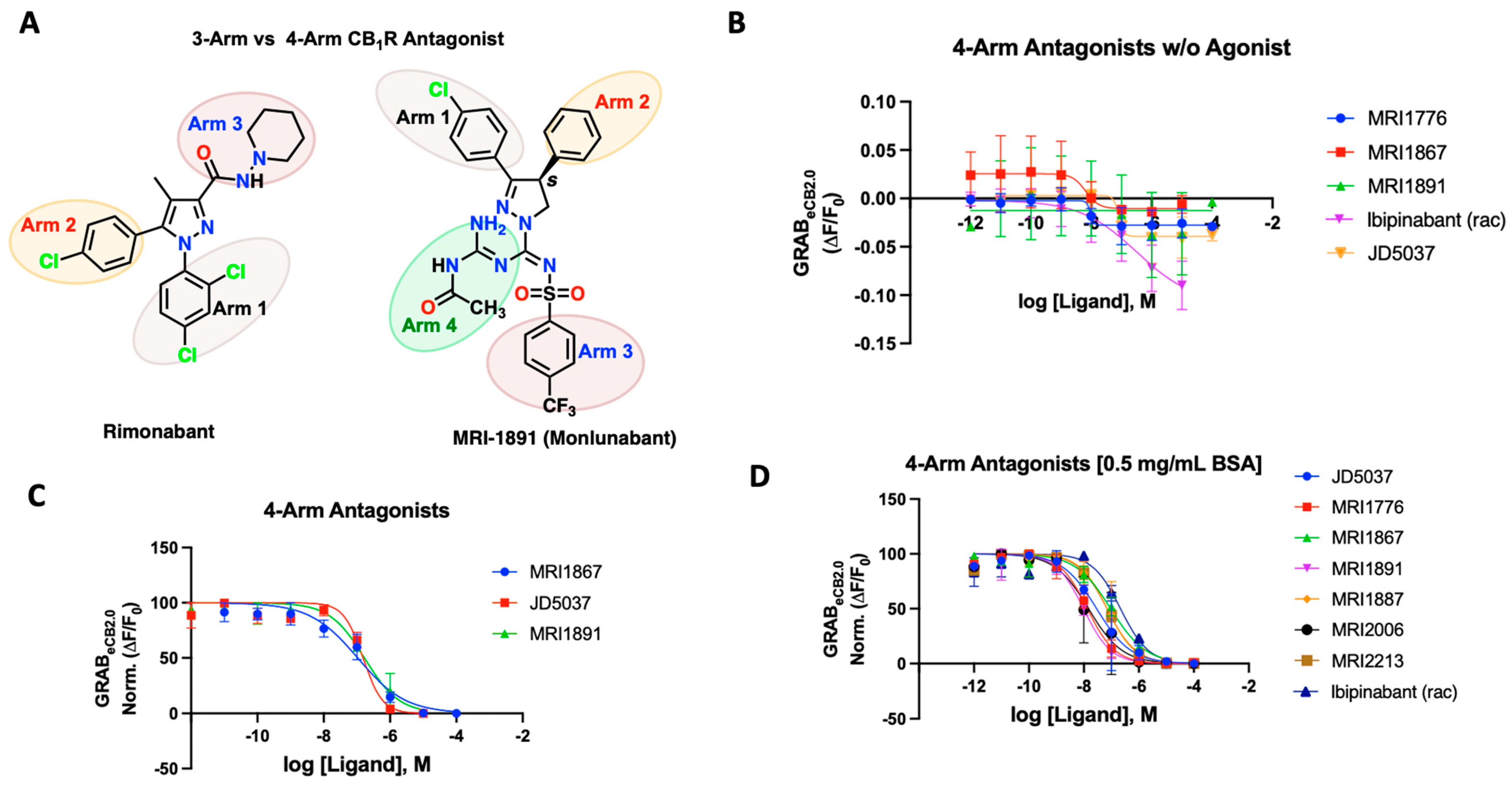
2.4. 4-Arm Modulators
2.5. GRABeCB2.0 Assays for Allosteric Modulators
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodríguez de Fonseca, F.; Del Arco, I.; Bermudez-Silva, F.J.; Bilbao, A.; Cippitelli, A.; Navarro, M. The endocannabinoid system: Physiology and pharmacology. Alcohol Alcohol. 2005, 40, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Howlett, A.C.; Abood, M.E. CB1 and CB2 receptor pharmacology. Adv. Pharmacol. 2017, 80, 169–206. [Google Scholar] [PubMed]
- Howlett, A.C.; Barth, F.; Bonner, T.I.; Cabral, G.; Casellas, P.; Devane, W.A.; Felder, C.C.; Herkenham, M.; Mackie, K.; Martin, B.R.; et al. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol. Rev. 2002, 54, 161–202. [Google Scholar] [CrossRef]
- Pacher, P.; Bátkai, S.; Kunos, G. The Endocannabinoid System as an Emerging Target of Pharmacotherapy. Pharmacol. Rev. 2006, 58, 389–462. [Google Scholar] [CrossRef]
- Galiegue, S.; Mary, S.; Marchand, J.; Dussossoy, D.; Carriere, D.; Carayon, P.; Bouaboula, M.; Shire, D.; Le Fur, G.; Casellas, P. Expression of Central and Peripheral Cannabinoid Receptors in Human Immune Tissues and Leukocyte Subpopulations. Eur. J. Biochem. 1995, 232, 54–61. [Google Scholar] [CrossRef]
- Onaivi, E.S.; Ishiguro, H.; Gong, J.P.; Patel, S.; Perchuk, A.; Meozzi, P.A.; Myers, L.; Mora, Z.; Tagliaferro, P.; Gardner, E.; et al. Discovery of the Presence and Functional Expression of Cannabinoid CB2 Receptors in Brain. Ann. N. Y. Acad. Sci. 2006, 1074, 514–536. [Google Scholar] [CrossRef]
- Cabral, G.A.; Raborn, E.S.; Griffin, L.; Dennis, J.; Marciano-Cabral, F. CB2 receptors in the brain: Role in central immune function. Br. J. Pharmacol. 2008, 153, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Turu, G.; Hunyady, L. Signal transduction of the CB1 cannabinoid receptor. J. Mol. Endocrinol. 2009, 44, 75–85. [Google Scholar] [CrossRef]
- Delgado-Peraza, F.; Ahn, K.H.; Nogueras-Ortiz, C.; Mungrue, I.N.; Mackie, K.; Kendall, D.A.; Yudowski, G.A. Mechanisms of Biased β-Arrestin-Mediated Signaling Downstream from the Cannabinoid 1 Receptor. Mol. Pharmacol. 2016, 89, 618–629. [Google Scholar] [CrossRef]
- Pacher, P.; Kunos, G. Modulating the endocannabinoid system in human health and disease—Successes and failures. FEBS J. 2013, 280, 1918–1943. [Google Scholar] [CrossRef]
- Sun, L.-J.; Yu, J.-W.; Wan, L.; Zhang, X.-Y.; Shi, Y.-G.; Chen, M.-Y. Endocannabinoid system activation contributes to glucose metabolism disorders of hepatocytes and promotes hepatitis C virus replication. Int. J. Infect. Dis. 2014, 23, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Gatta-Cherifi, B.; Cota, D. New insights on the role of the endocannabinoid system in the regulation of energy balance. Int. J. Obes. 2016, 40, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Han, J.H.; Kim, W. Peripheral CB1R as a modulator of metabolic inflammation. FASEB J. 2021, 35, e21232. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, M.; Bátkai, S.; Kechrid, M.; Mukhopadhyay, P.; Lee, W.-S.; Horváth, B.; Holovac, E.; Cinar, R.; Liaudet, L.; Mackie, K.; et al. Cannabinoid 1 receptor promotes cardiac dysfunction, oxidative stress, inflammation, and fibrosis in diabetic cardiomyopathy. Diabetes 2012, 61, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Cinar, R.; Iyer, M.R.; Kunos, G. The therapeutic potential of second and third generation CB1R antagonists. Pharmacol. Ther. 2020, 208, 107477. [Google Scholar] [CrossRef] [PubMed]
- Janero, D.R.; Makriyannis, A. Cannabinoid receptor antagonists: Pharmacological opportunities, clinical experience, and translational prognosis. Expert Opin. Emerg. Drugs 2009, 14, 43–65. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.J.; Raffa, R.B. Rimonabant Redux and Strategies to Improve the Future Outlook of CB1 Receptor Neutral-Antagonist/Inverse-Agonist Therapies. Obesity 2011, 19, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Pertwee, R.G. Cannabinoid receptor ligands: Clinical and neuropharmacological considerations, relevant to future drug discovery and development. Expert Opin. Investig. Drugs 2000, 9, 1553–1571. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.K.; Murumkar, P.R.; Kanhed, A.M.; Giridhar, R.; Yadav, M.R. Prospective therapeutic agents for obesity: Molecular modification approaches of centrally and peripherally acting selective cannabinoid 1 receptor antagonists. Eur. J. Med. Chem. 2014, 79, 298–339. [Google Scholar] [CrossRef]
- Dong, A.; He, K.; Dudok, B.; Farrell, J.S.; Guan, W.; Liput, D.J.; Puhl, H.L.; Cai, R.; Wang, H.; Duan, J.; et al. A fluorescent sensor for spatiotemporally resolved imaging of endocannabinoid dynamics in vivo. Nat. Biotechnol. 2022, 40, 787–798. [Google Scholar] [CrossRef]
- Singh, S.; Sarroza, D.; English, A.; McGrory, M.; Dong, A.; Zweifel, L.; Land, B.B.; Li, Y.; Bruchas, M.R.; Stella, N. Pharmacological Characterization of the Endocannabinoid Sensor GRABeCB2.0. Cannabis Cannabinoid Res. 2023. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.; Singh, S.; Hook, S.; Tyndall, J.D.A.; Vernall, A.J. Chemical Tools for Studying Lipid-Binding Class A G Protein–Coupled Receptors. Pharmacol. Rev. 2017, 69, 316–353. [Google Scholar] [CrossRef] [PubMed]
- Ravotto, L.; Duffet, L.; Zhou, X.; Weber, B.; Patriarchi, T. A Bright and Colorful Future for G-Protein Coupled Receptor Sensors. Front. Cell. Neurosci. 2020, 14, 67. [Google Scholar] [CrossRef] [PubMed]
- Haider, R.S.; Godbole, A.; Hoffmann, C. To sense or not to sense—New insights from GPCR-based and arrestin-based biosensors. Curr. Opin. Cell Biol. 2019, 57, 16–24. [Google Scholar] [CrossRef]
- Zhang, R.; Xie, X. Tools for GPCR drug discovery. Acta Pharmacol. Sin. 2012, 33, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.A.; Grieb, M.; Lutz, B. Central side-effects of therapies based on CB1 cannabinoid receptor agonists and antagonists: Focus on anxiety and depression. Best Pract. Res. Clin. Endocrinol. Metab. 2009, 23, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, D.L.; Abbott, J.K. Cannabinoids and the brain: The effects of endogenous and exogenous cannabinoids on brain systems and function. In The Complex Connection between Cannabis and Schizophrenia; Elsevier: Amsterdam, The Netherlands, 2018; pp. 37–74. [Google Scholar]
- Milligan, A.L.; Szabo-Pardi, T.A.; Burton, M.D. Cannabinoid receptor type 1 and its role as an analgesic: An opioid alternative? J. Dual Diagn. 2020, 16, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, J.; Saady, M.; Cardounel, A.; Kalimi, M. Identification and characterization of a novel synthetic cannabinoid CP 55,940 binder in rat brain cytosol. Mol. Cell. Biochem. 1998, 181, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi-Carmona, M.; Pialot, F.; Congy, C.; Redon, E.; Barth, F.; Bachy, A.; Brelière, J.-C.; Soubrié, P.; Le Fur, G. Characterization and distribution of binding sites for [3H]-SR 141716A, a selective brain (CB1) cannabinoid receptor antagonist, in rodent brain. Life Sci. 1996, 58, 1239–1247. [Google Scholar] [CrossRef]
- Darmani, N.A.; Sim-Selley, L.J.; Martin, B.R.; Janoyan, J.J.; Crim, J.L.; Parekh, B.; Breivogel, C.S. Antiemetic and motor-depressive actions of CP55,940: Cannabinoid CB1 receptor characterization, distribution, and G-protein activation. Eur. J. Pharmacol. 2003, 459, 83–95. [Google Scholar] [CrossRef]
- Gilbert, M.T.; Sulik, K.K.; Fish, E.W.; Baker, L.K.; Dehart, D.B.; Parnell, S.E. Dose-dependent teratogenicity of the synthetic cannabinoid CP-55,940 in mice. Neurotoxicol. Teratol. 2016, 58, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Fang, Y.-Q.; Ren, H.; Chen, T.; Guo, J.-J.; Yan, J.; Song, S.; Zhang, L.-Y.; Liao, H. WIN55,212-2 protects oligodendrocyte precursor cells in stroke penumbra following permanent focal cerebral ischemia in rats. Acta Pharmacol. Sin. 2013, 34, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Porcella, A.; Maxia, C.; Gessa, G.L.; Pani, L. The synthetic cannabinoid WIN55212-2 decreases the intraocular pressure in human glaucoma resistant to conventional therapies. Eur. J. Neurosci. 2001, 13, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Diego, M.; Angelina, A.; Martín-Cruz, L.; de la Rocha-Muñoz, A.; Maldonado, A.; Sevilla-Ortega, C.; Palomares, O. Cannabinoid WIN55,212-2 reprograms monocytes and macrophages to inhibit LPS-induced inflammation. Front. Immunol. 2023, 14, 1147520. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zeng, Y.; Zhou, Y.; Yu, J.; Liang, M.; Qin, L.; Zhou, Y. Win55,212-2 improves neural injury induced by HIV-1 glycoprotein 120 in rats by exciting CB2R. Brain Res. Bull. 2022, 182, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Herzberg, U.; Eliav, E.; Bennett, G.; Kopin, I.J. The analgesic effects of R(+)-WIN 55,212–2 mesylate, a high affinity cannabinoid agonist, in a rat model of neuropathic pain. Neurosci. Lett. 1997, 221, 157–160. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Peigneur, S.; Hendrickx, L.A.; Tytgat, J. Targeting Cannabinoid Receptors: Current Status and Prospects of Natural Products. Int. J. Mol. Sci. 2020, 21, 5064. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hua, T.; Vemuri, K.; Ho, J.H.; Wu, Y.; Wu, L.; Popov, P.; Benchama, O.; Zvonok, N.; Qu, L.; et al. Crystal structure of the human cannabinoid receptor CB2. Cell 2019, 176, 459–467.e13. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Thomas, B.F.; Zhang, Y. Overcoming the Psychiatric Side Effects of the Cannabinoid CB1 Receptor Antagonists: Current Approaches for Therapeutics Development. Curr. Top. Med. Chem. 2019, 19, 1418–1435. [Google Scholar] [CrossRef]
- Shao, Z.; Yin, J.; Chapman, K.; Grzemska, M.; Clark, L.; Wang, J.; Rosenbaum, D.M. High-resolution crystal structure of the human CB1 cannabinoid receptor. Nature 2016, 540, 602–606. [Google Scholar] [CrossRef]
- Patel, P.N.; Pathak, R. Rimonabant: A novel selective cannabinoid-1 receptor antagonist for treatment of obesity. Am. J. Health Pharm. 2007, 64, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.R.; Cinar, R.; Liu, J.; Godlewski, G.; Szanda, G.; Puhl, H.; Ikeda, S.R.; Deschamps, J.; Lee, Y.-S.; Steinbach, P.J.; et al. Structural Basis of Species-Dependent Differential Affinity of 6-Alkoxy-5-Aryl-3-Pyridinecarboxamide Cannabinoid-1 Receptor Antagonists. Mol. Pharmacol. 2015, 88, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, K.; Rosenbaum, D.M. Molecular basis for ligand modulation of the cannabinoid CB1 receptor. Br. J. Pharmacol. 2022, 179, 3487–3495. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.D.; Zhang, L.; Kavanagh, K.; Ward, G.M.; Chin, J.E.; Hadcock, J.R.; Auerbach, B.J.; Harwood, H.J. A Selective Cannabinoid-1 Receptor Antagonist, PF-95453, Reduces Body Weight and Body Fat to a Greater Extent than Pair-Fed Controls in Obese Monkeys. J. Pharmacol. Exp. Ther. 2010, 335, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Griffith, D.A.; Hadcock, J.R.; Black, S.C.; Iredale, P.A.; Carpino, P.A.; DaSilva-Jardine, P.; Day, R.; DiBrino, J.; Dow, R.L.; Landis, M.S.; et al. Discovery of 1-[9-(4-chlorophenyl)-8-(2-chlorophenyl)-9H-purin-6-yl]-4-ethylaminopiperidine-4-carboxylic acid amide hydrochloride (CP-945,598), a novel, potent, and selective cannabinoid type 1 receptor antagonist. J. Med. Chem. 2009, 52, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Chorvat, R.J. Peripherally restricted CB1 receptor blockers. Bioorg. Med. Chem. Lett. 2013, 23, 4751–4760. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.R.; Cinar, R.; Coffey, N.J.; Kunos, G. Synthesis of 13 C6-labeled, dual-target inhibitor of cannabinoid-1 receptor (CB1 R) and inducible nitric oxide synthase (iNOS). J. Labelled Comp. Radiopharm. 2018, 61, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Iyer, M.R.; Godlewski, G.; Jourdan, T.; Liu, J.; Coffey, N.J.; Zawatsky, C.N.; Puhl, H.L.; Wess, J.; Meister, J.; et al. Functional Selectivity of a Biased Cannabinoid-1 Receptor (CB1R) Antagonist. ACS Pharmacol. Transl. Sci. 2021, 4, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Cinar, R.; Iyer, M.R.; Liu, Z.; Cao, Z.; Jourdan, T.; Erdelyi, K.; Godlewski, G.; Szanda, G.; Liu, J.; Park, J.K.; et al. Hybrid inhibitor of peripheral cannabinoid-1 receptors and inducible nitric oxide synthase mitigates liver fibrosis. J. Clin. Investig. 2016, 1, e87336. [Google Scholar] [CrossRef]
- Iyer, M.R.; Cinar, R.; Katz, A.; Gao, M.; Erdelyi, K.; Jourdan, T.; Coffey, N.J.; Pacher, P.; Kunos, G. Design, Synthesis, and Biological Evaluation of Novel, Non-Brain-Penetrant, Hybrid Cannabinoid CB1R Inverse Agonist/Inducible Nitric Oxide Synthase (iNOS) Inhibitors for the Treatment of Liver Fibrosis. J. Med. Chem. 2017, 60, 1126–1141. [Google Scholar] [CrossRef]
- Iyer, M.R.; Cinar, R.; Wood, C.M.; Zawatsky, C.N.; Coffey, N.J.; Kim, K.A.; Liu, Z.; Katz, A.; Abdalla, J.; Hassan, S.A.; et al. Synthesis, Biological Evaluation, and Molecular Modeling Studies of 3,4-Diarylpyrazoline Series of Compounds as Potent, Nonbrain Penetrant Antagonists of Cannabinoid-1 (CB1R) Receptor with Reduced Lipophilicity. J. Med. Chem. 2022, 65, 2374–2387. [Google Scholar] [CrossRef] [PubMed]
- Dvorácskó, S.; Herrerias, A.; Oliverio, A.; Bhattacharjee, P.; Pommerolle, L.; Liu, Z.; Feng, D.; Lee, Y.-S.; Hassan, S.A.; Godlewski, G.; et al. Cannabinoformins: Designing Biguanide-Embedded, Orally Available, Peripherally Selective Cannabinoid-1 Receptor Antagonists for Metabolic Syndrome Disorders. J. Med. Chem. 2023, 66, 11985–12004. [Google Scholar] [CrossRef] [PubMed]
- Santos-Molina, L.; Herrerias, A.; Zawatsky, C.N.; Gunduz-Cinar, O.; Cinar, R.; Iyer, M.R.; Wood, C.M.; Lin, Y.; Gao, B.; Kunos, G.; et al. Effects of a Peripherally Restricted Hybrid Inhibitor of CB1 Receptors and iNOS on Alcohol Drinking Behavior and Alcohol-Induced Endotoxemia. Molecules 2021, 26, 5089. [Google Scholar] [CrossRef] [PubMed]
- Roger, C.; Buch, C.; Muller, T.; Leemput, J.; Demizieux, L.; Passilly-Degrace, P.; Cinar, R.; Iyer, M.R.; Kunos, G.; Vergès, B.; et al. Simultaneous Inhibition of Peripheral CB1R and iNOS Mitigates Obesity-Related Dyslipidemia Through Distinct Mechanisms. Diabetes 2020, 69, 2120–2132. [Google Scholar] [CrossRef] [PubMed]
- Zawatsky, C.N.; Park, J.K.; Abdalla, J.; Kunos, G.; Iyer, M.R.; Cinar, R. Peripheral Hybrid CB1R and iNOS Antagonist MRI-1867 Displays Anti-Fibrotic Efficacy in Bleomycin-Induced Skin Fibrosis. Front. Endocrinol. 2021, 12, 744857. [Google Scholar] [CrossRef]
- Cinar, R.; Park, J.K.; Zawatsky, C.N.; Coffey, N.J.; Bodine, S.P.; Abdalla, J.; Yokoyama, T.; Jourdan, T.; Jay, L.; Zuo, M.X.G.; et al. CB1R and iNOS are distinct players promoting pulmonary fibrosis in Hermansky–Pudlak syndrome. Clin. Transl. Med. 2021, 11, e471. [Google Scholar] [CrossRef]
- Crater, G.D.; Lalonde, K.; Ravenelle, F.; Harvey, M.; Després, J.-P. Effects of CB1R inverse agonist, INV-202, in patients with features of metabolic syndrome. A randomized, placebo-controlled, double-blind phase 1b study. Diabetes Obes. Metab. 2023, 26, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Chorvat, R.J.; Berbaum, J.; Seriacki, K.; McElroy, J.F. JD-5006 and JD-5037: Peripherally restricted (PR) cannabinoid-1 receptor blockers related to SLV-319 (Ibipinabant) as metabolic disorder therapeutics devoid of CNS liabilities. Bioorg. Med. Chem. Lett. 2012, 22, 6173–6180. [Google Scholar] [CrossRef]
- Lange, J.H.; Coolen, H.K.; van Stuivenberg, H.H.; Dijksman, J.A.; Herremans, A.H.; Ronken, E.; Keizer, H.G.; Tipker, K.; McCreary, A.C.; Veerman, W.; et al. Synthesis, biological properties, and molecular modeling investigations of novel 3,4-diarylpyrazolines as potent and selective CB(1) cannabinoid receptor antagonists. J. Med. Chem. 2004, 47, 627–643. [Google Scholar] [CrossRef]
- Janero, D.R.; Thakur, G.A. Leveraging allostery to improve G protein-coupled receptor (GPCR)-directed therapeutics: Cannabinoid receptor 1 as discovery target. Expert Opin. Drug Discov. 2016, 11, 1223–1237. [Google Scholar] [CrossRef]
- Ross, R.A. Allosterism and cannabinoid CB1 receptors: The shape of things to come. Trends Pharmacol. Sci. 2007, 28, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.T.; Vinson, P.N.; Niswender, C.M. Approaches for probing allosteric interactions at 7 transmembrane spanning receptors. Prog. Mol. Biol. Transl. Sci. 2013, 115, 1–59. [Google Scholar] [PubMed]
- Morales, P.; Goya, P.; Jagerovic, N.; Hernandez-Folgado, L. Allosteric Modulators of the CB1 Cannabinoid Receptor: A Structural Update Review. Cannabis Cannabinoid Res. 2016, 1, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Dopart, R.; Lu, D.; Lichtman, A.H.; Kendall, D.A. Allosteric modulators of cannabinoid receptor 1: Developing compounds for improved specificity. Drug Metab. Rev. 2018, 50, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Price, M.R.; Baillie, G.L.; Thomas, A.; Stevenson, L.A.; Easson, M.; Goodwin, R.; McLean, A.; McIntosh, L.; Goodwin, G.; Walker, G.; et al. Allosteric Modulation of the Cannabinoid CB1 Receptor. Mol. Pharmacol. 2005, 68, 1484–1495. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K.H.; Mahmoud, M.M.; Kendall, D.A. Allosteric Modulator ORG27569 Induces CB1 Cannabinoid Receptor High Affinity Agonist Binding State, Receptor Internalization, and Gi Protein-independent ERK1/2 Kinase Activation. J. Biol. Chem. 2012, 287, 12070–12082. [Google Scholar] [CrossRef]
- Horswill, J.G.; Bali, U.; Shaaban, S.; Keily, J.F.; Jeevaratnam, P.; Babbs, A.J.; Reynet, C.; Wong Kai In, P. PSNCBAM-1, a novel allosteric antagonist at cannabinoid CB1 receptors with hypophagic effects in rats. Br. J. Pharmacol. 2007, 152, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Arunlakshana, O.; Schild, H.O. Some quantitative uses of drug antagonists. Br. J. Pharmacol. Chemother. 1959, 14, 48–58. [Google Scholar] [CrossRef]
- Laprairie, R.B.; Kulkarni, P.M.; Deschamps, J.R.; Kelly, M.E.M.; Janero, D.R.; Cascio, M.G.; Stevenson, L.A.; Pertwee, R.G.; Kenakin, T.P.; Denovan-Wright, E.M.; et al. Enantiospecific Allosteric Modulation of Cannabinoid 1 Receptor. ACS Chem. Neurosci. 2017, 8, 1188–1203. [Google Scholar] [CrossRef]
- Thomas, M.; Lu, B.; Breschi, J.J.; Ge, Q.; Zhang, C.; Chen, J.; Klibanov, A.M. Fulldeacylation of polyethylenimine dramatically boosts itsgene delivery efficiency and specificity to mouse lung. Proc. Natl. Acad. Sci. USA 2005, 102, 5679–5684. [Google Scholar]
- Calderone, V.; Baragatti, B.; Breschi, M.; Martinotti, E. Experimental and theoretical comparisons between the classical Schild analysis and a new alternative method to evaluate the pA2 of competitive antagonists. Naunyn-Schmiedeberg’s Arch. Pharmacol. 1999, 360, 477–487. [Google Scholar] [CrossRef]
- Lew, M.; Angus, J. Analysis of competitive agonist-antagonist interactions by nonlinear regression. Trends Pharmacol. Sci. 1995, 16, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.S.; Lanza, T.J.; Jewell, J.P.; Liu, P.; Shah, S.K.; Qi, H.; Tong, X.; Wang, J.; Xu, S.S.; Fong, T.M.; et al. Discovery of N-[(1S,2S)-3-(4-Chlorophenyl)-2- (3-Cyanophenyl)-1-Methylpropyl]-2-Methyl-2- {[5-(Trifluoromethyl)Pyridin-2-Yl]Oxy}propanamide (MK-0364), a Novel, Acyclic Cannabinoid-1 Receptor Inverse Agonist for the Treatment of Obesity. J. Med. Chem. 2006, 49, 7584–7587. [Google Scholar] [CrossRef] [PubMed]




| Compound | GRABeCB2.0 IC50 [nM] | GRABeCB2.0 IC50 [nM] 0.5 mg/mL BSA |
|---|---|---|
| Rimonabant | 31 ± 9.5 | 25 ± 2.5 |
| Taranabant | 205 ± 15 | 23 ± 5 |
| Otenabant | 27 ± 7.5 | 41 ± 4.8 |
| Compound | GRABeCB2.0 IC50 [nM] | GRABeCB2.0 IC50 [nM] 0.5 mg/mL BSA |
|---|---|---|
| MRI-1867 | 105 ± 5.4 | 90 ± 5 |
| MRI-1891 | 168 ± 57 | 9.8 ± 1.4 |
| JD5037 | 148 ± 12 | 29 ± 3.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shivshankar, S.; Nimely, J.; Puhl, H., III; Iyer, M.R. Pharmacological Evaluation of Cannabinoid Receptor Modulators Using GRABeCB2.0 Sensor. Int. J. Mol. Sci. 2024, 25, 5012. https://doi.org/10.3390/ijms25095012
Shivshankar S, Nimely J, Puhl H III, Iyer MR. Pharmacological Evaluation of Cannabinoid Receptor Modulators Using GRABeCB2.0 Sensor. International Journal of Molecular Sciences. 2024; 25(9):5012. https://doi.org/10.3390/ijms25095012
Chicago/Turabian StyleShivshankar, Samay, Josephine Nimely, Henry Puhl, III, and Malliga R. Iyer. 2024. "Pharmacological Evaluation of Cannabinoid Receptor Modulators Using GRABeCB2.0 Sensor" International Journal of Molecular Sciences 25, no. 9: 5012. https://doi.org/10.3390/ijms25095012






