Recent Developments in Autism Genetic Research: A Scientometric Review from 2018 to 2022
Abstract
:1. Introduction
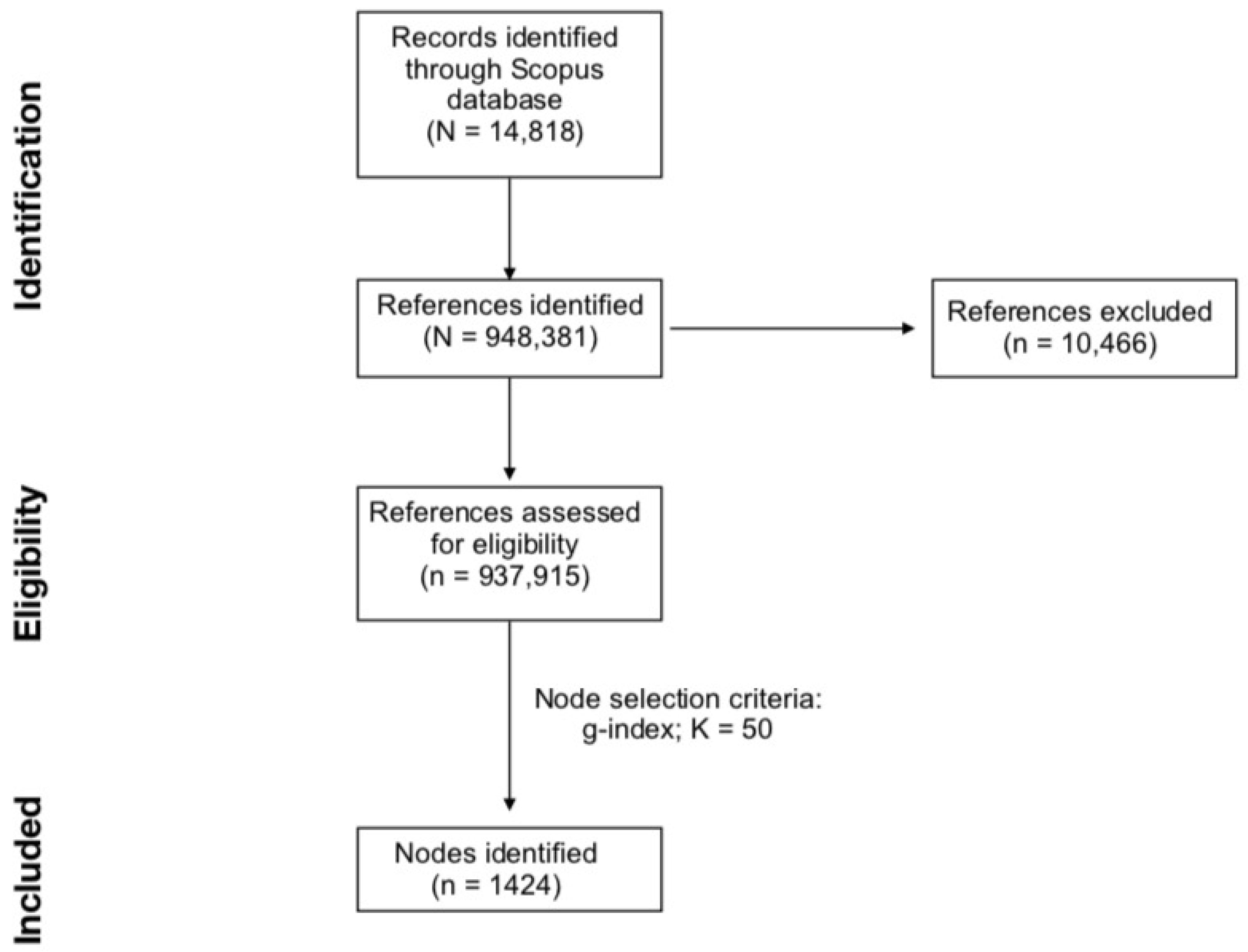
2. Materials and Methods
2.1. Data Collection and Conversion
2.2. Document Co-Citation Analysis
2.3. Metrics
3. Results
3.1. Structural Properties of DCA Network
3.2. Documents with a Citation Burst
4. Discussion
4.1. Cluster #0: Networks and Pathways
4.2. Cluster #1: Gut Microbiota
4.3. Clusters #2 and #3: Mouse Models
4.4. Clusters #4 and #6: Stem Cell Technology
4.5. Cluster #5: Genomic Architecture
4.6. Cluster #7: Psychiatric Disorder
4.7. Cluster #8: Sex Difference
4.8. Cluster #9: Copy Number Variations (CNVs)
4.9. Cluster #10: Developmental Perspectives
4.10. Cluster #14: Antiseizure Drug
4.11. Limitations and Future Recommendations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ASD | Autism Spectrum Disorder |
| DCA | Document Co-Citation Analysis |
| LLR | Log-Likelihood Ratio |
| GCS | Global Citing Score |
| ILAE | International League Against Epilepsy |
References
- Rosanoff, A.J. A Theory of Personality Based Mainly on Psychiatric Experience. Psychol. Bull. 1920, 17, 281. [Google Scholar] [CrossRef]
- Kanner, L. Early infantile autism. J. Pediatr. 1944, 25, 211–217. [Google Scholar] [CrossRef]
- Volkmar, F.R.; Bregman, J.; Cohen, D.J.; Cicchetti, D.V. DSM-III and DSM-III-R diagnoses of autism. Am. J. Psychiatry 1988, 145, 1404–1408. [Google Scholar] [PubMed]
- Grzadzinski, R.; Huerta, M.; Lord, C. DSM-5 and Autism Spectrum Disorders (ASDs): An opportunity for identifying ASD subtypes. Mol. Autism 2013, 4, 12. [Google Scholar] [CrossRef]
- Chiarotti, F.; Venerosi, A. Epidemiology of Autism Spectrum Disorders: A review of worldwide prevalence estimates since 2014. Brain Sci. 2020, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Fombonne, E. Is there an epidemic of autism? Pediatrics 2001, 107, 411–412. [Google Scholar] [CrossRef]
- Randall, M.; Sciberras, E.; Brignell, A.; Ihsen, E.; Efron, D.; Dissanayake, C.; Williams, K. Autism Spectrum Disorder: Presentation and prevalence in a nationally representative Australian sample. Aust. N. Z. J. Psychiatry 2016, 50, 243–253. [Google Scholar] [CrossRef]
- Ouellette-Kuntz, H.; Coo, H.; Lam, M.; Breitenbach, M.M.; Hennessey, P.E.; Jackman, P.D.; Lewis, M.; Dewey, D.; Bernier, F.P.; Chung, A.M. The changing prevalence of autism in three regions of Canada. J. Autism Dev. Disord. 2014, 44, 120–136. [Google Scholar] [CrossRef]
- MAY, J.; Dunn, L. A preliminary study of inherited blood group antigens in families having an index case of early infantile autism. J. Hered. 1961, 52, 239–240. [Google Scholar] [CrossRef]
- Gayon, J. From Mendel to epigenetics: History of genetics. Comptes Rendus Biol. 2016, 339, 225–230. [Google Scholar] [CrossRef] [Green Version]
- Muhle, R.; Trentacoste, S.V.; Rapin, I. The genetics of autism. Pediatrics 2004, 113, e472–e486. [Google Scholar] [CrossRef] [PubMed]
- Geschwind, D.H. Genetics of Autism Spectrum Disorders. Trends Cogn. Sci. 2011, 15, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Qiu, Y.; Li, Y.; Cong, X. Genetics of Autism Spectrum Disorder: An umbrella review of systematic reviews and meta-analyses. Transl. Psychiatry 2022, 12, 249. [Google Scholar] [CrossRef]
- Rylaarsdam, L.; Guemez-Gamboa, A. Genetic causes and modifiers of Autism Spectrum Disorder. Front. Cell. Neurosci. 2019, 13, 385. [Google Scholar] [CrossRef] [PubMed]
- Cataldo, I.; Azhari, A.; Esposito, G. A review of oxytocin and arginine-vasopressin receptors and their modulation of Autism Spectrum Disorder. Front. Mol. Neurosci. 2018, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Azhari, A.; Azizan, F.; Esposito, G. A systematic review of gut-immune-brain mechanisms in Autism Spectrum Disorder. Dev. Psychobiol. 2019, 61, 752–771. [Google Scholar] [CrossRef]
- Jahan, N.; Naveed, S.; Zeshan, M.; Tahir, M.A. How to conduct a systematic review: A narrative literature review. Cureus 2016, 8, e864. [Google Scholar] [CrossRef]
- Mulchenko, Z. Measurement of science. study of the development of science as an information process. In USAF Foreign Technology Division Translation AD735634; National Technical Information Service: Springfield, VA, USA, 1969. [Google Scholar]
- Börner, K.; Chen, C.; Boyack, K.W. Visualizing knowledge domains. Annu. Rev. Inf. Sci. Technol. 2003, 37, 179–255. [Google Scholar] [CrossRef]
- Junior, O.F. The Legacy of David Bohm in Physics—An Essay in Scientometry. In David Bohm; Springer: Cham, Switzerland, 2019; pp. 223–240. [Google Scholar]
- Su, H.N.; Lee, P.C. Mapping knowledge structure by keyword co-occurrence: A first look at journal papers in Technology Foresight. Scientometrics 2010, 85, 65–79. [Google Scholar] [CrossRef]
- Cataldo, I.; Lieu, A.A.; Carollo, A.; Bornstein, M.H.; Gabrieli, G.; Lee, A.; Esposito, G. From the cradle to the web: The growth of “sharenting”—A scientometric perspective. Hum. Behav. Emerg. Technol. 2022, 2022, 5607422. [Google Scholar] [CrossRef]
- Aryadoust, V.; Zakaria, A.; Lim, M.H.; Chen, C. An extensive knowledge mapping review of measurement and validity in language assessment and SLA research. Front. Psychol. 2020, 11, 1941. [Google Scholar] [CrossRef] [PubMed]
- Chen, C. Science mapping: A systematic review of the literature. J. Data Inf. Sci. 2017, 2, 1–40. [Google Scholar] [CrossRef]
- Chen, C. CiteSpace II: Detecting and visualizing emerging trends and transient patterns in scientific literature. J. Am. Soc. Inf. Sci. Technol. 2006, 57, 359–377. [Google Scholar] [CrossRef]
- Chen, C. CiteSpace: A Practical Guide for Mapping Scientific Literature; Nova Science Publishers: Hauppauge, NY, USA, 2016. [Google Scholar]
- Small, H. Co-citation context analysis and the structure of paradigms. J. Doc. 1980, 36, 183–196. [Google Scholar] [CrossRef]
- Chen, C.; Ibekwe-SanJuan, F.; Hou, J. The structure and dynamics of cocitation clusters: A multiple-perspective cocitation analysis. J. Am. Soc. Inf. Sci. Technol. 2010, 61, 1386–1409. [Google Scholar] [CrossRef]
- Egghe, L. An improvement of the h-index: The G-index. ISSI Newsl. 2006, 2, 8–9. [Google Scholar]
- Bornmann, L.; Daniel, H.D. What do we know about the h index? J. Am. Soc. Inf. Sci. Technol. 2007, 58, 1381–1385. [Google Scholar] [CrossRef]
- Alonso, S.; Cabrerizo, F.J.; Herrera-Viedma, E.; Herrera, F. h-Index: A review focused in its variants, computation and standardization for different scientific fields. J. Inf. 2009, 3, 273–289. [Google Scholar] [CrossRef]
- Chen, C. The citespace manual. Coll. Comput. Inform. 2014, 1, 1–84. [Google Scholar]
- Carollo, A.; Balagtas, J.P.M.; Neoh, M.J.Y.; Esposito, G. A scientometric approach to review the role of the medial preoptic area (MPOA) in parental behavior. Brain Sci. 2021, 11, 393. [Google Scholar] [CrossRef]
- Newman, M.E. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef] [PubMed]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Aryadoust, V.; Ang, B.H. Exploring the frontiers of eye tracking research in language studies: A novel co-citation scientometric review. Comput. Assist. Lang. Learn. 2019, 34, 898–933. [Google Scholar] [CrossRef]
- Freeman, L.C. A set of measures of centrality based on betweenness. Sociometry 1977, 40, 35–41. [Google Scholar] [CrossRef]
- Kleinberg, J. Bursty and hierarchical structure in streams. Data Min. Knowl. Discov. 2003, 7, 373–397. [Google Scholar] [CrossRef]
- Gaggero, G.; Bonassi, A.; Dellantonio, S.; Pastore, L.; Aryadoust, V.; Esposito, G. A scientometric review of alexithymia: Mapping thematic and disciplinary shifts in half a century of research. Front. Psychiatry 2020, 11, 1405. [Google Scholar] [CrossRef]
- Carollo, A.; Lim, M.; Aryadoust, V.; Esposito, G. Interpersonal Synchrony in the Context of Caregiver-Child Interactions: A Document Co-citation Analysis. Front. Psychol. 2021, 12, 2977. [Google Scholar] [CrossRef]
- Lord, C.; Elsabbagh, M.; Baird, G.; Veenstra-Vanderweele, J. Autism Spectrum Disorder. Lancet 2018, 392, 508–520. [Google Scholar] [CrossRef]
- Grove, J.; Ripke, S.; Als, T.D.; Mattheisen, M.; Walters, R.K.; Won, H.; Pallesen, J.; Agerbo, E.; Andreassen, O.A.; Anney, R.; et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019, 51, 431–444. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Muotri, A.R.; Sebat, J. Getting to the cores of autism. Cell 2019, 178, 1287–1298. [Google Scholar] [CrossRef]
- Lim, M.; Carollo, A.; Chen, S.A.; Esposito, G. Surveying 80 years of psychodrama research: A scientometric review. Front. Psychiatry 2021, 12, 780542. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Cruz, N.J.; Kang, D.W.; Gandal, M.J.; Wang, B.; Kim, Y.M.; Zink, E.M.; Casey, C.P.; Taylor, B.C.; Lane, C.J.; et al. Human gut microbiota from Autism Spectrum Disorder promote behavioral symptoms in mice. Cell 2019, 177, 1600–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruzzo, E.K.; Pérez-Cano, L.; Jung, J.Y.; Wang, L.k.; Kashef-Haghighi, D.; Hartl, C.; Singh, C.; Xu, J.; Hoekstra, J.N.; Leventhal, O.; et al. Inherited and de novo genetic risk for autism impacts shared networks. Cell 2019, 178, 850–866. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Leventhal, B.L.; Koh, Y.J.; Fombonne, E.; Laska, E.; Lim, E.C.; Cheon, K.A.; Kim, S.J.; Kim, Y.K.; Lee, H.; et al. Prevalence of Autism Spectrum Disorders in a total population sample. Am. J. Psychiatry 2011, 168, 904–912. [Google Scholar] [CrossRef]
- Abraham, A.; Milham, M.P.; Di Martino, A.; Craddock, R.C.; Samaras, D.; Thirion, B.; Varoquaux, G. Deriving reproducible biomarkers from multi-site resting-state data: An Autism-based example. NeuroImage 2017, 147, 736–745. [Google Scholar] [CrossRef]
- Lim, E.T.; Uddin, M.; De Rubeis, S.; Chan, Y.; Kamumbu, A.S.; Zhang, X.; D’Gama, A.M.; Kim, S.N.; Hill, R.S.; Goldberg, A.P.; et al. Rates, distribution and implications of postzygotic mosaic mutations in Autism Spectrum Disorder. Nat. Neurosci. 2017, 20, 1217–1224. [Google Scholar] [CrossRef]
- Yang, M.; Bozdagi, O.; Scattoni, M.L.; Wöhr, M.; Roullet, F.I.; Katz, A.M.; Abrams, D.N.; Kalikhman, D.; Simon, H.; Woldeyohannes, L.; et al. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. J. Neurosci. 2012, 32, 6525–6541. [Google Scholar] [CrossRef]
- Lee, P.H.; Anttila, V.; Won, H.; Feng, Y.C.A.; Rosenthal, J.; Zhu, Z.; Tucker-Drob, E.M.; Nivard, M.G.; Grotzinger, A.D.; Posthuma, D.; et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 2019, 179, 1469–1482. [Google Scholar] [CrossRef]
- Nowakowski, T.J.; Bhaduri, A.; Pollen, A.A.; Alvarado, B.; Mostajo-Radji, M.A.; Di Lullo, E.; Haeussler, M.; Sandoval-Espinosa, C.; Liu, S.J.; Velmeshev, D.; et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 2017, 358, 1318–1323. [Google Scholar] [CrossRef]
- Velmeshev, D.; Schirmer, L.; Jung, D.; Haeussler, M.; Perez, Y.; Mayer, S.; Bhaduri, A.; Goyal, N.; Rowitch, D.H.; Kriegstein, A.R. Single-cell genomics identifies cell type–specific molecular changes in autism. Science 2019, 364, 685–689. [Google Scholar] [CrossRef]
- Matta, S.M.; Hill-Yardin, E.L.; Crack, P.J. The influence of neuroinflammation in Autism Spectrum Disorder. Brain Behav. Immun. 2019, 79, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Estes, M.L.; McAllister, A.K. Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat. Rev. Neurosci. 2015, 16, 469–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goines, P.E.; Ashwood, P. Cytokine dysregulation in Autism Spectrum Disorders (ASD): Possible role of the environment. Neurotoxicol. Teratol. 2013, 36, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Pantelis, C.; Papadimitriou, G.N.; Papiol, S.; Parkhomenko, E.; Pato, M.T.; Paunio, T.; Pejovic-Milovancevic, M.; Perkins, D.O.; Pietiläinen, O.; Pimm, J. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar]
- Yuen, R.K.; Thiruvahindrapuram, B.; Merico, D.; Walker, S.; Tammimies, K.; Hoang, N.; Chrysler, C.; Nalpathamkalam, T.; Pellecchia, G.; Liu, Y.; et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med. 2015, 21, 185–191. [Google Scholar] [CrossRef]
- Antoine, M.W.; Langberg, T.; Schnepel, P.; Feldman, D.E. Increased excitation-inhibition ratio stabilizes synapse and circuit excitability in four autism mouse models. Neuron 2019, 101, 648–661. [Google Scholar] [CrossRef]
- Schafer, S.T.; Paquola, A.; Stern, S.; Gosselin, D.; Ku, M.; Pena, M.; Kuret, T.J.; Liyanage, M.; Mansour, A.A.; Jaeger, B.N.; et al. Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nat. Neurosci. 2019, 22, 243–255. [Google Scholar] [CrossRef]
- Stahl, E.A.; Breen, G.; Forstner, A.J.; McQuillin, A.; Ripke, S.; Trubetskoy, V.; Mattheisen, M.; Wang, Y.; Coleman, J.R.; Gaspar, H.A.; et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 2019, 51, 793–803. [Google Scholar] [CrossRef]
- Joensuu, M.; Lanoue, V.; Hotulainen, P. Dendritic spine actin cytoskeleton in Autism Spectrum Disorder. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 84, 362–381. [Google Scholar] [CrossRef]
- Alonso-Gonzalez, A.; Rodriguez-Fontenla, C.; Carracedo, A. De novo mutations (DNMs) in Autism Spectrum Disorder (ASD): Pathway and network analysis. Front. Genet. 2018, 9, 406. [Google Scholar] [CrossRef]
- Ayhan, F.; Konopka, G. Regulatory genes and pathways disrupted in Autism Spectrum Disorders. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2019, 89, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.; Katayama, Y.; Nakayama, K.I.; Nomura, J.; Sakurai, T. Characterizing vulnerable brain areas and circuits in mouse models of autism: Towards understanding pathogenesis and new therapeutic approaches. Neurosci. Biobehav. Rev. 2020, 110, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Eyring, K.W.; Geschwind, D.H. Three decades of ASD genetics: Building a foundation for neurobiological understanding and treatment. Hum. Mol. Genet. 2021, 30, R236–R244. [Google Scholar] [CrossRef]
- Gandhi, T.; Lee, C.C. Neural mechanisms underlying repetitive behaviors in rodent models of Autism Spectrum Disorders. Front. Cell. Neurosci. 2021, 14, 592710. [Google Scholar] [CrossRef] [PubMed]
- Voineagu, I.; Wang, X.; Johnston, P.; Lowe, J.K.; Tian, Y.; Horvath, S.; Mill, J.; Cantor, R.M.; Blencowe, B.J.; Geschwind, D.H. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 2011, 474, 380–384. [Google Scholar] [CrossRef]
- Parikshak, N.N.; Luo, R.; Zhang, A.; Won, H.; Lowe, J.K.; Chandran, V.; Horvath, S.; Geschwind, D.H. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 2013, 155, 1008–1021. [Google Scholar] [CrossRef]
- De Rubeis, S.; He, X.; Goldberg, A.P.; Poultney, C.S.; Samocha, K.; Ercument Cicek, A.; Kou, Y.; Liu, L.; Fromer, M.; Walker, S.; et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014, 515, 209–215. [Google Scholar] [CrossRef]
- Guang, S.; Pang, N.; Deng, X.; Yang, L.; He, F.; Wu, L.; Chen, C.; Yin, F.; Peng, J. Synaptopathology involved in Autism Spectrum Disorder. Front. Cell. Neurosci. 2018, 12, 470. [Google Scholar] [CrossRef]
- Garcia-Forn, M.; Boitnott, A.; Akpinar, Z.; De Rubeis, S. Linking autism risk genes to disruption of cortical development. Cells 2020, 9, 2500. [Google Scholar] [CrossRef]
- Diaz-Caneja, C.M.; State, M.; Hagerman, R.; Jacquemont, S.; Marín, O.; Bagni, C.; Umbricht, D.; Simonoff, E.; de Andrés-Trelles, F.; Kaale, A.; et al. A white paper on a neurodevelopmental framework for drug discovery in autism and other neurodevelopmental disorders. Eur. Neuropsychopharmacol. 2021, 48, 49–88. [Google Scholar] [CrossRef]
- DiCarlo, G.E.; Wallace, M.T. Modeling dopamine dysfunction in Autism Spectrum Disorder: From invertebrates to vertebrates. Neurosci. Biobehav. Rev. 2021, 133, 104494. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Verhoeff, T.A.; Perez Pardo, P.; Garssen, J.; Kraneveld, A.D. The gut–brain axis in Autism Spectrum Disorder: A focus on the metalloproteases ADAM10 and ADAM17. Int. J. Mol. Sci. 2020, 22, 118. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.; Sandhu, K.V.; Bastiaanssen, T.F.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The microbiota-gut–brain axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Fattorusso, A.; Di Genova, L.; Dell’Isola, G.B.; Mencaroni, E.; Esposito, S. Autism Spectrum Disorders and the gut microbiota. Nutrients 2019, 11, 521. [Google Scholar] [CrossRef]
- Srikantha, P.; Mohajeri, M.H. The possible role of the microbiota-gut–brain-axis in autism spectrum disorder. Int. J. Mol. Sci. 2019, 20, 2115. [Google Scholar] [CrossRef] [PubMed]
- Hooks, K.B.; Konsman, J.P.; O’Malley, M.A. Microbiota-gut–brain research: A critical analysis. Behav. Brain Sci. 2019, 42, e60. [Google Scholar] [CrossRef] [PubMed]
- Azhari, A.; Azizan, F.; Esposito, G. Beyond a gut feeling: How the immune system impacts the effect of gut microbiota in neurodevelopment. Behav. Brain Sci. 2019, 42. [Google Scholar] [CrossRef]
- van Sadelhoff, J.H.; Perez Pardo, P.; Wu, J.; Garssen, J.; Van Bergenhenegouwen, J.; Hogenkamp, A.; Hartog, A.; Kraneveld, A.D. The gut-immune-brain axis in Autism Spectrum Disorders; a focus on amino acids. Front. Endocrinol. 2019, 10, 247. [Google Scholar] [CrossRef]
- Tomova, A.; Husarova, V.; Lakatosova, S.; Bakos, J.; Vlkova, B.; Babinska, K.; Ostatnikova, D. Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav. 2015, 138, 179–187. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, X.; Dan, Z.; Pei, Y.; Xu, R.; Guo, M.; Liu, K.; Zhang, F.; Chen, J.; Su, C.; et al. Gene variations in Autism Spectrum Disorder are associated with alternation of gut microbiota, metabolites and cytokines. Gut Microbes 2021, 13, 1854967. [Google Scholar] [CrossRef]
- Taniya, M.A.; Chung, H.J.; Al Mamun, A.; Alam, S.; Aziz, M.; Emon, N.U.; Islam, M.; Podder, B.R.; Ara Mimi, A.; Aktar Suchi, S.; et al. Role of Gut Microbiome in Autism Spectrum Disorder and Its Therapeutic Regulation. Front. Cell. Infect. Microbiol. 2022, 12, 915701. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Lukkes, J.L.; Shekhar, A. Overview of genetic models of Autism Spectrum Disorders. Prog. Brain Res. 2018, 241, 1–36. [Google Scholar] [PubMed]
- Yang, G.; Shcheglovitov, A. Probing disrupted neurodevelopment in autism using human stem cell-derived neurons and organoids: An outlook into future diagnostics and drug development. Dev. Dyn. 2020, 249, 6–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panisi, C.; Guerini, F.R.; Abruzzo, P.M.; Balzola, F.; Biava, P.M.; Bolotta, A.; Brunero, M.; Burgio, E.; Chiara, A.; Clerici, M.; et al. Autism Spectrum Disorder from the womb to adulthood: Suggestions for a paradigm shift. J. Pers. Med. 2021, 11, 70. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, M.X.; Sun, L.; Wallis, C.U.; Zhou, J.S.; Ao, L.J.; Li, Q.; Sham, P.C. Rational use of mesenchymal stem cells in the treatment of autism spectrum disorders. World J. Stem Cells 2019, 11, 55. [Google Scholar] [CrossRef]
- Lombardo, M.V.; Moon, H.M.; Su, J.; Palmer, T.D.; Courchesne, E.; Pramparo, T. Maternal immune activation dysregulation of the fetal brain transcriptome and relevance to the pathophysiology of autism spectrum disorder. Mol. Psychiatry 2018, 23, 1001–1013. [Google Scholar] [CrossRef]
- Möhrle, D.; Fernández, M.; Peñagarikano, O.; Frick, A.; Allman, B.; Schmid, S. What we can learn from a genetic rodent model about autism. Neurosci. Biobehav. Rev. 2020, 109, 29–53. [Google Scholar] [CrossRef]
- Verma, V.; Paul, A.; Amrapali Vishwanath, A.; Vaidya, B.; Clement, J.P. Understanding intellectual disability and Autism Spectrum Disorders from common mouse models: Synapses to behaviour. Open Biol. 2019, 9, 180265. [Google Scholar] [CrossRef]
- Nakai, N.; Takumi, T.; Nakai, J.; Sato, M. Common defects of spine dynamics and circuit function in neurodevelopmental disorders: A systematic review of findings from in vivo optical imaging of mouse models. Front. Neurosci. 2018, 12, 412. [Google Scholar] [CrossRef]
- Sungur, A.Ö.; Schwarting, R.K.; Woehr, M. Behavioral phenotypes and neurobiological mechanisms in the Shank1 mouse model for Autism Spectrum Disorder: A translational perspective. Behav. Brain Res. 2018, 352, 46–61. [Google Scholar] [CrossRef]
- Wang, X.; Kery, R.; Xiong, Q. Synaptopathology in Autism Spectrum Disorders: Complex effects of synaptic genes on neural circuits. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 84, 398–415. [Google Scholar] [CrossRef] [PubMed]
- Soler, J.; Fañanás, L.; Parellada, M.; Krebs, M.O.; Rouleau, G.A.; Fatjó-Vilas, M. Genetic variability in scaffolding proteins and risk for schizophrenia and autism-spectrum disorders: A systematic review. J. Psychiatry Neurosci. 2018, 43, 223–244. [Google Scholar] [CrossRef] [PubMed]
- Consorthium, T.D.B.F.X.; Bakker, C.E.; Verheij, C.; Willemsen, R.; van der Helm, R.; Oerlemans, F.; Vermey, M.; Bygrave, A.; Hoogeveen, A.; Oostra, B.A.; et al. Fmr1 knockout mice: A model to study fragile X mental retardation. Cell 1994, 78, 23–33. [Google Scholar]
- Bagni, C.; Zukin, R.S. A synaptic perspective of fragile X syndrome and autism spectrum disorders. Neuron 2019, 101, 1070–1088. [Google Scholar] [CrossRef] [Green Version]
- Kaufmann, W.E.; Kidd, S.A.; Andrews, H.F.; Budimirovic, D.B.; Esler, A.; Haas-Givler, B.; Stackhouse, T.; Riley, C.; Peacock, G.; Sherman, S.L.; et al. Autism Spectrum Disorder in fragile X syndrome: Cooccurring conditions and current treatment. Pediatrics 2017, 139, S194–S206. [Google Scholar] [CrossRef]
- Clifford, S.; Dissanayake, C.; Bui, Q.M.; Huggins, R.; Taylor, A.K.; Loesch, D.Z. Autism spectrum phenotype in males and females with fragile X full mutation and premutation. J. Autism Dev. Disord. 2007, 37, 738–747. [Google Scholar] [CrossRef]
- Verkerk, A.J.; Pieretti, M.; Sutcliffe, J.S.; Fu, Y.H.; Kuhl, D.P.; Pizzuti, A.; Reiner, O.; Richards, S.; Victoria, M.F.; Zhang, F.; et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 1991, 65, 905–914. [Google Scholar] [CrossRef]
- Gibson, J.R.; Bartley, A.F.; Hays, S.A.; Huber, K.M. Imbalance of neocortical excitation and inhibition and altered UP states reflect network hyperexcitability in the mouse model of fragile X syndrome. J. Neurophysiol. 2008, 100, 2615–2626. [Google Scholar] [CrossRef]
- Yoo, T.; Cho, H.; Park, H.; Lee, J.; Kim, E. Shank3 exons 14–16 deletion in glutamatergic neurons leads to social and repetitive behavioral deficits associated with increased cortical layer 2/3 neuronal excitability. Front. Cell. Neurosci. 2019, 13, 458. [Google Scholar] [CrossRef]
- Mossa, A.; Giona, F.; Pagano, J.; Sala, C.; Verpelli, C. SHANK genes in autism: Defining therapeutic targets. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 84, 416–423. [Google Scholar] [CrossRef]
- Yoo, T.; Cho, H.; Lee, J.; Park, H.; Yoo, Y.E.; Yang, E.; Kim, J.Y.; Kim, H.; Kim, E. GABA neuronal deletion of Shank3 exons 14–16 in mice suppresses striatal excitatory synaptic input and induces social and locomotor abnormalities. Front. Cell. Neurosci. 2018, 12, 341. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, P.; Feng, G. SHANK proteins: Roles at the synapse and in Autism Spectrum Disorder. Nat. Rev. Neurosci. 2017, 18, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Chaudry, S.; Vasudevan, N. mTOR-dependent spine dynamics in autism. Front. Mol. Neurosci. 2022, 15, 877609. [Google Scholar] [CrossRef]
- Ali Rodriguez, R.; Joya, C.; Hines, R.M. Common ribs of inhibitory synaptic dysfunction in the umbrella of neurodevelopmental disorders. Front. Mol. Neurosci. 2018, 11, 132. [Google Scholar] [CrossRef] [Green Version]
- St. Clair, D.; Johnstone, M. Using mouse transgenic and human stem cell technologies to model genetic mutations associated with schizophrenia and autism. Philos. Trans. R. Soc. Biol. Sci. 2018, 373, 20170037. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Brugha, T.S.; Charman, T.; Cusack, J.; Dumas, G.; Frazier, T.; Jones, E.J.; Jones, R.M.; Pickles, A.; State, M.W.; et al. Autism Spectrum Disorder. Nat. Rev. Dis. Prim. 2020, 6, 1–23. [Google Scholar] [CrossRef]
- Vallortigara, G. Born Knowing: Imprinting and the Origins of Knowledge; MIT Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Tartaglione, A.M.; Schiavi, S.; Calamandrei, G.; Trezza, V. Prenatal valproate in rodents as a tool to understand the neural underpinnings of social dysfunctions in Autism Spectrum Disorder. Neuropharmacology 2019, 159, 107477. [Google Scholar] [CrossRef]
- Fink, J.J.; Levine, E.S. Uncovering true cellular phenotypes: Using induced pluripotent stem cell-derived neurons to study early insults in neurodevelopmental disorders. Front. Neurol. 2018, 9, 237. [Google Scholar] [CrossRef]
- Maussion, G.; Rocha, C.; Bernard, G.; Beitel, L.K.; Durcan, T.M. Patient-derived stem cells, another in vitro model, or the missing link toward novel therapies for Autism Spectrum Disorders? Front. Pediatr. 2019, 7, 225. [Google Scholar] [CrossRef]
- Sato, A.; Kotajima-Murakami, H.; Tanaka, M.; Katoh, Y.; Ikeda, K. Influence of prenatal drug exposure, maternal inflammation, and parental aging on the development of Autism Spectrum Disorder. Front. Psychiatry 2022, 13, 821455. [Google Scholar] [CrossRef]
- Ilieva, M.; Svenningsen, Å.F.; Thorsen, M.; Michel, T.M. Psychiatry in a dish: Stem cells and brain organoids modeling autism spectrum disorders. Biol. Psychiatry 2018, 83, 558–568. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.K.; Griffiths, R.; Price, D.J.; Mason, J.O. Cerebral organoids as tools to identify the developmental roots of autism. Mol. Autism 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Baldassari, S.; Musante, I.; Iacomino, M.; Zara, F.; Salpietro, V.; Scudieri, P. Brain organoids as model systems for genetic neurodevelopmental disorders. Front. Cell Dev. Biol. 2020, 8, 590119. [Google Scholar] [CrossRef] [PubMed]
- Shou, Y.; Liang, F.; Xu, S.; Li, X. The application of brain organoids: From neuronal development to neurological diseases. Front. Cell Dev. Biol. 2020, 8, 579659. [Google Scholar] [CrossRef]
- de Jong, J.O.; Llapashtica, C.; Genestine, M.; Strauss, K.; Provenzano, F.; Sun, Y.; Zhu, H.; Cortese, G.P.; Brundu, F.; Brigatti, K.W.; et al. Cortical overgrowth in a preclinical forebrain organoid model of CNTNAP2-associated Autism Spectrum Disorder. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Sun, N.; Meng, X.; Liu, Y.; Song, D.; Jiang, C.; Cai, J. Applications of brain organoids in neurodevelopment and neurological diseases. J. Biomed. Sci. 2021, 28, 1–16. [Google Scholar] [CrossRef]
- Silverman, J.L.; Thurm, A.; Ethridge, S.B.; Soller, M.M.; Petkova, S.P.; Abel, T.; Bauman, M.D.; Brodkin, E.S.; Harony-Nicolas, H.; Wöhr, M.; et al. Reconsidering animal models used to study Autism Spectrum Disorder: Current state and optimizing future. Genes Brain Behav. 2022, 21, e12803. [Google Scholar] [CrossRef]
- Tordjman, S.; Drapier, D.; Bonnot, O.; Graignic, R.; Fortes, S.; Cohen, D.; Millet, B.; Laurent, C.; Roubertoux, P.L. Animal models relevant to schizophrenia and autism: Validity and limitations. Behav. Genet. 2007, 37, 61–78. [Google Scholar] [CrossRef]
- Siniscalco, D.; Sapone, A.; Cirillo, A.; Giordano, C.; Maione, S.; Antonucci, N. Autism Spectrum Disorders: Is mesenchymal stem cell personalized therapy the future? J. Biomed. Biotechnol. 2012, 2012, 480289. [Google Scholar] [CrossRef]
- Filice, F.; Janickova, L.; Henzi, T.; Bilella, A.; Schwaller, B. The parvalbumin hypothesis of Autism Spectrum Disorder. Front. Cell. Neurosci. 2020, 14, 577525. [Google Scholar] [CrossRef]
- Napolitano, A.; Schiavi, S.; La Rosa, P.; Rossi-Espagnet, M.C.; Petrillo, S.; Bottino, F.; Tagliente, E.; Longo, D.; Lupi, E.; Casula, L.; et al. Sex Differences in Autism Spectrum Disorder: Diagnostic, Neurobiological, and Behavioral Features. Front. Psychiatry 2022, 13, 889636. [Google Scholar] [CrossRef] [PubMed]
- Courchesne, E.; Gazestani, V.H.; Lewis, N.E. Prenatal origins of ASD: The when, what, and how of ASD development. Trends Neurosci. 2020, 43, 326–342. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Ziller, M.; Spengler, D. Childhood-onset schizophrenia: Insights from induced pluripotent stem cells. Int. J. Mol. Sci. 2018, 19, 3829. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.J.; Vogelstein, J.T.; Gozzi, A.; Bernhardt, B.C.; Yeo, B.T.; Milham, M.P.; Di Martino, A. Toward neurosubtypes in autism. Biol. Psychiatry 2020, 88, 111–128. [Google Scholar] [CrossRef] [Green Version]
- Niu, W.; Parent, J.M. Modeling genetic epilepsies in a dish. Dev. Dyn. 2020, 249, 56–75. [Google Scholar] [CrossRef]
- Fetit, R.; Hillary, R.F.; Price, D.J.; Lawrie, S.M. The neuropathology of autism: A systematic review of post-mortem studies of autism and related disorders. Neurosci. Biobehav. Rev. 2021, 129, 35–62. [Google Scholar] [CrossRef]
- Griesi-Oliveira, K.; Fogo, M.; Pinto, B.; Alves, A.; Suzuki, A.; Morales, A.; Ezquina, S.; Sosa, O.; Sutton, G.; Sunaga-Franze, D.; et al. Transcriptome of iPSC-derived neuronal cells reveals a module of co-expressed genes consistently associated with Autism Spectrum Disorder. Mol. Psychiatry 2021, 26, 1589–1605. [Google Scholar] [CrossRef]
- Al-Dewik, N.; Al-Jurf, R.; Styles, M.; Tahtamouni, S.; Alsharshani, D.; Alsharshani, M.; Ahmad, A.I.; Khattab, A.; Al Rifai, H.; Walid Qoronfleh, M. Overview and introduction to Autism Spectrum Disorder (ASD). In Personalized Food Intervention and Therapy for Autism Spectrum Disorder Management; Springer: Cham, Switzerland, 2020; pp. 3–42. [Google Scholar]
- Breen, M.S.; Browne, A.; Hoffman, G.E.; Stathopoulos, S.; Brennand, K.; Buxbaum, J.D.; Drapeau, E. Transcriptional signatures of participant-derived neural progenitor cells and neurons implicate altered Wnt signaling in Phelan-McDermid syndrome and autism. Mol. Autism 2020, 11, 53. [Google Scholar] [CrossRef]
- Seabra, C.M.; Aneichyk, T.; Erdin, S.; Tai, D.J.; De Esch, C.E.; Razaz, P.; An, Y.; Manavalan, P.; Ragavendran, A.; Stortchevoi, A.; et al. Transcriptional consequences of MBD5 disruption in mouse brain and CRISPR-derived neurons. Mol. Autism 2020, 11, 45. [Google Scholar] [CrossRef]
- Brusco, A.; Ferrero, G.B. Genomic Architecture of ASD. In Psychopathology in Adolescents and Adults with Autism Spectrum Disorders; Springer: Cham, Switzerland, 2019; pp. 23–34. [Google Scholar]
- Rogozin, I.B.; Gertz, E.M.; Baranov, P.V.; Poliakov, E.; Schaffer, A.A. Genome-wide changes in protein translation efficiency are associated with autism. Genome Biol. Evol. 2018, 10, 1902–1919. [Google Scholar] [CrossRef]
- Chen, T.; Froehlich, T.; Li, T.; Lu, L. Big data approaches to develop a comprehensive and accurate tool aimed at improving Autism Spectrum Disorder diagnosis and subtype stratification. Libr. Tech. 2020, 38, 819–833. [Google Scholar] [CrossRef]
- Nehme, R.; Barrett, L.E. Using human pluripotent stem cell models to study autism in the era of big data. Mol. Autism 2020, 11, 21. [Google Scholar] [CrossRef]
- Liu, H.; Barnes, J.; Pedrosa, E.; Herman, N.S.; Salas, F.; Wang, P.; Zheng, D.; Lachman, H.M. Transcriptome analysis of neural progenitor cells derived from Lowe syndrome induced pluripotent stem cells: Identification of candidate genes for the neurodevelopmental and eye manifestations. J. Neurodev. Disord. 2020, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Cantor, R.M.; Lowe, J.K. Genome-wide association studies and neurodevelopment: Autism spectrum disorders and related traits. In Factors Affecting Neurodevelopment; Elsevier: Cham, Switzerland, 2021; pp. 27–37. [Google Scholar]
- Culotta, L.; Penzes, P. Exploring the mechanisms underlying excitation/inhibition imbalance in human iPSC-derived models of ASD. Mol. Autism 2020, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Gordon, A.; Geschwind, D.H. Human in vitro models for understanding mechanisms of autism spectrum disorder. Mol. Autism 2020, 11, 26. [Google Scholar] [CrossRef]
- Prem, S.; Millonig, J.H.; DiCicco-Bloom, E. Dysregulation of neurite outgrowth and cell migration in autism and other neurodevelopmental disorders. In Neurodevelopmental Disorders; Springer: Cham, Switzerland, 2020; pp. 109–153. [Google Scholar]
- Muhle, R.A.; Reed, H.E.; Stratigos, K.A.; Veenstra-VanderWeele, J. The emerging clinical neuroscience of Autism Spectrum Disorder: A review. JAMA Psychiatry 2018, 75, 514–523. [Google Scholar] [CrossRef]
- Grabrucker, A. Biometals in Autism Spectrum Disorders; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Saxena, R.; Babadi, M.; Namvarhaghighi, H.; Roullet, F.I. Role of environmental factors and epigenetics in autism spectrum disorders. Prog. Mol. Biol. Transl. Sci. 2020, 173, 35–60. [Google Scholar]
- Scuderi, C.; Verkhratsky, A. The role of neuroglia in Autism Spectrum Disorders. Prog. Mol. Biol. Transl. Sci. 2020, 173, 301–330. [Google Scholar]
- Zhang, X.; Abdellaoui, A.; Rucker, J.; de Jong, S.; Potash, J.B.; Weissman, M.M.; Shi, J.; Knowles, J.A.; Pato, C.; Pato, M.; et al. Genome-wide burden of rare short deletions is enriched in major depressive disorder in four cohorts. Biol. Psychiatry 2019, 85, 1065–1073. [Google Scholar] [CrossRef]
- Campbell, P.D.; Granato, M. Zebrafish as a tool to study schizophrenia-associated copy number variants. Dis. Model. Mech. 2020, 13, dmm043877. [Google Scholar] [CrossRef]
- Nehme, R.; Pietiläinen, O.; Artomov, M.; Tegtmeyer, M.; Valakh, V.; Lehtonen, L.; Bell, C.; Singh, T.; Trehan, A.; Sherwood, J.; et al. The 22q11. 2 region regulates presynaptic gene-products linked to schizophrenia. Nat. Commun. 2022, 13, 3690. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, F. Update on the genetic architecture of schizophrenia. Med. Genet. 2020, 32, 19–24. [Google Scholar] [CrossRef]
- Strom, N.I.; Grove, J.; Meier, S.M.; Bækvad-Hansen, M.; Nissen, J.B.; Als, T.D.; Halvorsen, M.; Nordentoft, M.; Mortensen, P.B.; Hougaard, D.M.; et al. Polygenic heterogeneity across obsessive-compulsive disorder subgroups defined by a comorbid diagnosis. Front. Genet. 2021, 12, 711624. [Google Scholar] [CrossRef] [PubMed]
- Bryois, J.; Skene, N.G.; Hansen, T.F.; Kogelman, L.J.; Watson, H.J.; Liu, Z.; Brueggeman, L.; Breen, G.; Bulik, C.M.; Arenas, E.; et al. Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson’s disease. Nat. Genet. 2020, 52, 482–493. [Google Scholar] [CrossRef]
- Yang, Z.; Wu, H.; Lee, P.H.; Tsetsos, F.; Davis, L.K.; Yu, D.; Lee, S.H.; Dalsgaard, S.; Haavik, J.; Barta, C.; et al. Investigating shared genetic basis across tourette syndrome and comorbid neurodevelopmental disorders along the impulsivity-compulsivity spectrum. Biol. Psychiatry 2021, 90, 317–327. [Google Scholar] [CrossRef]
- Warrier, V.; Baron-Cohen, S. Childhood trauma, life-time self-harm, and suicidal behaviour and ideation are associated with polygenic scores for autism. Mol. Psychiatry 2021, 26, 1670–1684. [Google Scholar] [CrossRef] [Green Version]
- Bralten, J.; Mota, N.R.; Klemann, C.J.; De Witte, W.; Laing, E.; Collier, D.A.; de Kluiver, H.; Bauduin, S.E.; Arango, C.; Ayuso-Mateos, J.L.; et al. Genetic underpinnings of sociability in the general population. Neuropsychopharmacology 2021, 46, 1627–1634. [Google Scholar] [CrossRef]
- Park, C.Y.; Zhou, J.; Wong, A.K.; Chen, K.M.; Theesfeld, C.L.; Darnell, R.B.; Troyanskaya, O.G. Genome-wide landscape of RNA-binding protein target site dysregulation reveals a major impact on psychiatric disorder risk. Nat. Genet. 2021, 53, 166–173. [Google Scholar] [CrossRef]
- Jiang, C.C.; Lin, L.S.; Long, S.; Ke, X.Y.; Fukunaga, K.; Lu, Y.M.; Han, F. Signalling pathways in Autism Spectrum Disorder: Mechanisms and therapeutic implications. Signal Transduct. Target. Ther. 2022, 7, 1–36. [Google Scholar] [CrossRef]
- Urresti, J.; Zhang, P.; Moran-Losada, P.; Yu, N.K.; Negraes, P.D.; Trujillo, C.A.; Antaki, D.; Amar, M.; Chau, K.; Pramod, A.B.; et al. Cortical organoids model early brain development disrupted by 16p11. 2 copy number variants in autism. Mol. Psychiatry 2021, 26, 7560–7580. [Google Scholar] [CrossRef]
- Walker, R.L.; Ramaswami, G.; Hartl, C.; Mancuso, N.; Gandal, M.J.; De La Torre-Ubieta, L.; Pasaniuc, B.; Stein, J.L.; Geschwind, D.H. Genetic control of expression and splicing in developing human brain informs disease mechanisms. Cell 2019, 179, 750–771. [Google Scholar] [CrossRef] [PubMed]
- Willsey, H.R.; Willsey, A.J.; Wang, B.; State, M.W. Genomics, convergent neuroscience and progress in understanding Autism Spectrum Disorder. Nat. Rev. Neurosci. 2022, 23, 323–341. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, P.F.; Geschwind, D.H. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell 2019, 177, 162–183. [Google Scholar] [CrossRef] [PubMed]
- Rees, E.; Owen, M.J. Translating insights from neuropsychiatric genetics and genomics for precision psychiatry. Genome Med. 2020, 12, 1–16. [Google Scholar] [CrossRef]
- Mullins, N.; Forstner, A.J.; O’Connell, K.S.; Coombes, B.; Coleman, J.R.; Qiao, Z.; Als, T.D.; Bigdeli, T.B.; Børte, S.; Bryois, J.; et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 2021, 53, 817–829. [Google Scholar] [CrossRef]
- Rujeedawa, T.; Zaman, S.H. The Diagnosis and Management of Autism Spectrum Disorder (ASD) in Adult Females in the Presence or Absence of an Intellectual Disability. Int. J. Environ. Res. Public Health 2022, 19, 1315. [Google Scholar] [CrossRef]
- Ruigrok, A.N.; Lai, M.C. Sex/gender differences in neurology and psychiatry: Autism. Handb. Clin. Neurol. 2020, 175, 283–297. [Google Scholar]
- Lombardo, M.V.; Auyeung, B.; Pramparo, T.; Quartier, A.; Courraud, J.; Holt, R.J.; Waldman, J.; Ruigrok, A.N.; Mooney, N.; Bethlehem, R.A.; et al. Sex-specific impact of prenatal androgens on social brain default mode subsystems. Mol. Psychiatry 2020, 25, 2175–2188. [Google Scholar] [CrossRef]
- Nowak, S.; Jacquemont, S. The effects of sex on prevalence and mechanisms underlying neurodevelopmental disorders. In Handbook of Clinical Neurology; Elsevier: Cham, Switzerland, 2020; Volume 173, pp. 327–339. [Google Scholar]
- Kallitsounaki, A.; Williams, D.M. Autism Spectrum Disorder and Gender Dysphoria/Incongruence. A systematic Literature Review and Meta-Analysis. J. Autism Dev. Disord. 2022, 1–15. [Google Scholar] [CrossRef]
- Loomes, R.; Hull, L.; Mandy, W.P.L. What is the male-to-female ratio in Autism Spectrum Disorder? A systematic review and meta-analysis. J. Am. Acad. Child Adolesc. Psychiatry 2017, 56, 466–474. [Google Scholar] [CrossRef]
- Lai, M.C.; Anagnostou, E.; Wiznitzer, M.; Allison, C.; Baron-Cohen, S. Evidence-based support for autistic people across the lifespan: Maximising potential, minimising barriers, and optimising the person–environment fit. Lancet Neurol. 2020, 19, 434–451. [Google Scholar] [CrossRef]
- Müller, R.A.; Fishman, I. Brain connectivity and neuroimaging of social networks in autism. Trends Cogn. Sci. 2018, 22, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Wilson, H.A.; Creighton, C.; Scharfman, H.; Choleris, E.; MacLusky, N.J. Endocrine insights into the pathophysiology of autism spectrum disorder. Neuroscientist 2021, 27, 650–667. [Google Scholar] [CrossRef] [PubMed]
- Howes, O.D.; Rogdaki, M.; Findon, J.L.; Wichers, R.H.; Charman, T.; King, B.H.; Loth, E.; McAlonan, G.M.; McCracken, J.T.; Parr, J.R.; et al. Autism Spectrum Disorder: Consensus guidelines on assessment, treatment and research from the British Association for Psychopharmacology. J. Psychopharmacol. 2018, 32, 3–29. [Google Scholar] [CrossRef]
- Yuen, T.; Carter, M.T.; Szatmari, P.; Ungar, W.J. Cost-effectiveness of genome and exome sequencing in children diagnosed with Autism Spectrum Disorder. Appl. Health Econ. Health Policy 2018, 16, 481–493. [Google Scholar] [CrossRef]
- Jønch, A.E.; Douard, E.; Moreau, C.; Van Dijck, A.; Passeggeri, M.; Kooy, F.; Puechberty, J.; Campbell, C.; Sanlaville, D.; Lefroy, H.; et al. Estimating the effect size of the 15Q11. 2 BP1–BP2 deletion and its contribution to neurodevelopmental symptoms: Recommendations for practice. J. Med. Genet. 2019, 56, 701–710. [Google Scholar] [CrossRef]
- Rosenberg, L.E.; Rosenberg, D.D. Human Genes and Genomes: Science, Health, Society; Academic Press: Cambridge, MA, USA, 2012; Volume 2. [Google Scholar]
- Shishido, E.; Aleksic, B.; Ozaki, N. Copy-number variation in the pathogenesis of autism spectrum disorder. Psychiatry Clin. Neurosci. 2014, 68, 85–95. [Google Scholar] [CrossRef]
- Egolf, L.E.; Vaksman, Z.; Lopez, G.; Rokita, J.L.; Modi, A.; Basta, P.V.; Hakonarson, H.; Olshan, A.F.; Diskin, S.J. Germline 16p11. 2 microdeletion predisposes to neuroblastoma. Am. J. Hum. Genet. 2019, 105, 658–668. [Google Scholar] [CrossRef]
- Lengyel, A.; Pinti, É.; Pikó, H.; Jávorszky, E.; David, D.; Tihanyi, M.; Gönczi, É.; Kiss, E.; Tóth, Z.; Tory, K.; et al. Clinical and genetic findings in Hungarian pediatric patients carrying chromosome 16p copy number variants and a review of the literature. Eur. J. Med. Genet. 2020, 63, 104027. [Google Scholar] [CrossRef]
- Bristow, G.C.; Thomson, D.M.; Openshaw, R.L.; Mitchell, E.J.; Pratt, J.A.; Dawson, N.; Morris, B.J. 16p11 Duplication disrupts hippocampal-orbitofrontal-amygdala connectivity, revealing a neural circuit endophenotype for schizophrenia. Cell Rep. 2020, 31, 107536. [Google Scholar] [CrossRef]
- Pucilowska, J.; Vithayathil, J.; Pagani, M.; Kelly, C.; Karlo, J.C.; Robol, C.; Morella, I.; Gozzi, A.; Brambilla, R.; Landreth, G.E. Pharmacological inhibition of ERK signaling rescues pathophysiology and behavioral phenotype associated with 16p11. 2 chromosomal deletion in mice. J. Neurosci. 2018, 38, 6640–6652. [Google Scholar] [CrossRef] [PubMed]
- Niarchou, M.; Chawner, S.J.; Doherty, J.L.; Maillard, A.M.; Jacquemont, S.; Chung, W.K.; Green-Snyder, L.; Bernier, R.A.; Goin-Kochel, R.P.; Hanson, E.; et al. Psychiatric disorders in children with 16p11. 2 deletion and duplication. Transl. Psychiatry 2019, 9, 1–8. [Google Scholar]
- Ip, J.P.; Nagakura, I.; Petravicz, J.; Li, K.; Wiemer, E.A.; Sur, M. Major vault protein, a candidate gene in 16p11. 2 microdeletion syndrome, is required for the homeostatic regulation of visual cortical plasticity. J. Neurosci. 2018, 38, 3890–3900. [Google Scholar] [CrossRef] [PubMed]
- Poot, M. Syndromes hidden within the 16p11. 2 deletion region. Mol. Syndromol. 2018, 9, 171–174. [Google Scholar] [CrossRef]
- Rein, B.; Yan, Z. 16p11. 2 copy number variations and neurodevelopmental disorders. Trends Neurosci. 2020, 43, 886–901. [Google Scholar] [CrossRef]
- Weiss, L.A.; Shen, Y.; Korn, J.M.; Arking, D.E.; Miller, D.T.; Fossdal, R.; Saemundsen, E.; Stefansson, H.; Ferreira, M.A.; Green, T.; et al. Association between microdeletion and microduplication at 16p11. 2 and autism. N. Engl. J. Med. 2008, 358, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.R.; Howrigan, D.P.; Merico, D.; Thiruvahindrapuram, B.; Wu, W.; Greer, D.S.; Antaki, D.; Shetty, A.; Holmans, P.A.; Pinto, D.; et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017, 49, 27–35. [Google Scholar] [CrossRef]
- Stefansson, H.; Meyer-Lindenberg, A.; Steinberg, S.; Magnusdottir, B.; Morgen, K.; Arnarsdottir, S.; Bjornsdottir, G.; Walters, G.B.; Jonsdottir, G.A.; Doyle, O.M.; et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature 2014, 505, 361–366. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, A.; Weiss, L.A. Recurrent reciprocal copy number variants: Roles and rules in neurodevelopmental disorders. Dev. Neurobiol. 2018, 78, 519–530. [Google Scholar] [CrossRef]
- Takumi, T.; Tamada, K. CNV biology in neurodevelopmental disorders. Curr. Opin. Neurobiol. 2018, 48, 183–192. [Google Scholar] [CrossRef]
- Kushima, I.; Aleksic, B.; Nakatochi, M.; Shimamura, T.; Okada, T.; Uno, Y.; Morikawa, M.; Ishizuka, K.; Shiino, T.; Kimura, H.; et al. Comparative analyses of copy-number variation in autism spectrum disorder and schizophrenia reveal etiological overlap and biological insights. Cell Rep. 2018, 24, 2838–2856. [Google Scholar] [CrossRef] [PubMed]
- Zwaigenbaum, L.; Duku, E.; Fombonne, E.; Szatmari, P.; Smith, I.M.; Bryson, S.E.; Mirenda, P.; Vaillancourt, T.; Volden, J.; Georgiades, S.; et al. Developmental functioning and symptom severity influence age of diagnosis in Canadian preschool children with autism. Paediatr. Child Health 2019, 24, e57–e65. [Google Scholar] [CrossRef] [PubMed]
- Atherton, G.; Edisbury, E.; Piovesan, A.; Cross, L. Autism through the ages: A mixed methods approach to understanding how age and age of diagnosis affect quality of life. J. Autism Dev. Disord. 2022, 52, 3639–3654. [Google Scholar] [CrossRef]
- Pender, R.; Fearon, P.; Heron, J.; Mandy, W. The longitudinal heterogeneity of autistic traits: A systematic review. Res. Autism Spectr. Disord. 2020, 79, 101671. [Google Scholar] [CrossRef]
- Kirst, S.; Diehm, R.; Bögl, K.; Wilde-Etzold, S.; Bach, C.; Noterdaeme, M.; Poustka, L.; Ziegler, M.; Dziobek, I. Fostering socio-emotional competencies in children on the autism spectrum using a parent-assisted serious game: A multicenter randomized controlled trial. Behav. Res. Ther. 2022, 152, 104068. [Google Scholar] [CrossRef] [PubMed]
- Rozenblatt-Perkal, Y.; Zaidman-Zait, A. Mother–child interaction in families of children with autism: Interpersonal dyadic processes. Res. Autism Spectr. Disord. 2020, 79, 101689. [Google Scholar] [CrossRef]
- Reinhartsen, D.; Tapia, A.; Watson, L.; Crais, E.; Bradley, C.; Fairchild, J.; Herring, A.; Daniels, J. Expressive dominant versus receptive dominant language patterns in young children: Findings from the study to explore early development. J. Autism Dev. Disord. 2019, 49, 2447–2460. [Google Scholar] [CrossRef]
- Belcher, H.L.; Morein-Zamir, S.; Stagg, S.D.; Ford, R.M. Shining a Light on a Hidden Population: Social Functioning and Mental Health in Women Reporting Autistic Traits But Lacking Diagnosis. J. Autism Dev. Disord. 2022, 1–15. [Google Scholar] [CrossRef]
- White, R.; Livingston, L.A.; Taylor, E.C.; Close, S.A.; Shah, P.; Callan, M.J. Understanding the contributions of trait autism and anxiety to extreme demand avoidance in the adult general population. J. Autism Dev. Disord. 2022, 1–9. [Google Scholar] [CrossRef]
- Brown, C.M.; Stokes, M.A. Intersection of eating disorders and the female profile of autism. Psychiatr. Clin. 2020, 43, 735–743. [Google Scholar]
- Simonoff, E.; Pickles, A.; Charman, T.; Chandler, S.; Loucas, T.; Baird, G. Psychiatric disorders in children with Autism Spectrum Disorders: Prevalence, comorbidity, and associated factors in a population-derived sample. J. Am. Acad. Child Adolesc. Psychiatry 2008, 47, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Barros, F.; Figueiredo, C.; Costa, A.; Soares, S.C. Sensory processing in the Autism Spectrum: The role of attention to detail and somatic trait anxiety in the olfactory perception of the general population. J. Autism Dev. Disord. 2021, 51, 2338–2353. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, A.; Nalborczyk, L.; Dutheil, F.; Kovarski, K.; Chokron, S.; Garrido, M.; Gomot, M.; Mermillod, M. High spatial frequency filtered primes hastens happy faces categorization in autistic adults. Brain Cogn. 2021, 155, 105811. [Google Scholar] [CrossRef] [PubMed]
- Hollin, G. From the profound to the mundane: Questionnaires as emerging technologies in autism genetics. Sci. Technol. Hum. Values 2019, 44, 634–659. [Google Scholar] [CrossRef]
- Nebel, M.B.; Lidstone, D.E.; Wang, L.; Benkeser, D.; Mostofsky, S.H.; Risk, B.B. Accounting for motion in resting-state fMRI: What part of the spectrum are we characterizing in Autism Spectrum Disorder? NeuroImage 2022, 257, 119296. [Google Scholar] [CrossRef]
- McCracken, J.T.; Anagnostou, E.; Arango, C.; Dawson, G.; Farchione, T.; Mantua, V.; McPartland, J.; Murphy, D.; Pandina, G.; Veenstra-VanderWeele, J.; et al. Drug development for Autism Spectrum Disorder (ASD): Progress, challenges, and future directions. Eur. Neuropsychopharmacol. 2021, 48, 3–31. [Google Scholar] [CrossRef]
- Hakami, T. Efficacy and tolerability of antiseizure drugs. Ther. Adv. Neurol. Disord. 2021, 14, 17562864211037430. [Google Scholar] [CrossRef]
- Hakami, T. Neuropharmacology of antiseizure drugs. Neuropsychopharmacol. Rep. 2021, 41, 336–351. [Google Scholar] [CrossRef]
- Frye, R.E.; Casanova, M.F.; Fatemi, S.H.; Folsom, T.D.; Reutiman, T.J.; Brown, G.L.; Edelson, S.M.; Slattery, J.C.; Adams, J.B. Neuropathological mechanisms of seizures in Autism Spectrum Disorder. Front. Neurosci. 2016, 10, 192. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, N.A.; Misra, S.N.; Jurado, M.; Kang, S.K.; Vierra, N.C.; Nguyen, K.; Wren, L.; George, A.L., Jr.; Trimmer, J.S.; Kearney, J.A. Epilepsy and neurobehavioral abnormalities in mice with a dominant-negative KCNB1 pathogenic variant. Neurobiol. Dis. 2021, 147, 105141. [Google Scholar] [CrossRef]
- De Maria, B.; Balestrini, S.; Mei, D.; Melani, F.; Pellacani, S.; Pisano, T.; Rosati, A.; Scaturro, G.M.; Giordano, L.; Cantalupo, G.; et al. Expanding the genetic and phenotypic spectrum of CHD2-related disease: From early neurodevelopmental disorders to adult-onset epilepsy. Am. J. Med. Genet. Part A 2022, 188, 522–533. [Google Scholar] [CrossRef]
- Cali, E.; Rocca, C.; Salpietro, V.; Houlden, H. Epileptic Phenotypes Associated with SNAREs and Related Synaptic Vesicle Exocytosis Machinery. Front. Neurol. 2021, 12, 806506. [Google Scholar] [CrossRef] [PubMed]
- Galanopoulou, A.S.; Löscher, W.; Lubbers, L.; O’Brien, T.J.; Staley, K.; Vezzani, A.; D’Ambrosio, R.; White, H.S.; Sontheimer, H.; Wolf, J.A.; et al. Antiepileptogenesis and disease modification: Progress, challenges, and the path forward—Report of the Preclinical Working Group of the 2018 NINDS-sponsored antiepileptogenesis and disease modification workshop. Epilepsia Open 2021, 6, 276–296. [Google Scholar] [CrossRef] [PubMed]
- Raga, S.; Specchio, N.; Rheims, S.; Wilmshurst, J.M. Developmental and epileptic encephalopathies: Recognition and approaches to care. Epileptic Disord. 2021, 23, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Scheffer, I.E.; Berkovic, S.; Capovilla, G.; Connolly, M.B.; French, J.; Guilhoto, L.; Hirsch, E.; Jain, S.; Mathern, G.W.; Moshé, S.L.; et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017, 58, 512–521. [Google Scholar] [CrossRef]
- Fisher, R.S.; Cross, J.H.; French, J.A.; Higurashi, N.; Hirsch, E.; Jansen, F.E.; Lagae, L.; Moshé, S.L.; Peltola, J.; Roulet Perez, E.; et al. Operational classification of seizure types by the International League Against Epilepsy: Position Paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017, 58, 522–530. [Google Scholar] [CrossRef]
- Fisher, R.S.; Acevedo, C.; Arzimanoglou, A.; Bogacz, A.; Cross, J.H.; Elger, C.E.; Engel, J., Jr.; Forsgren, L.; French, J.A.; Glynn, M.; et al. ILAE official report: A practical clinical definition of epilepsy. Epilepsia 2014, 55, 475–482. [Google Scholar] [CrossRef]
- Stamberger, H.; Hammer, T.B.; Gardella, E.; Vlaskamp, D.R.; Bertelsen, B.; Mandelstam, S.; de Lange, I.; Zhang, J.; Myers, C.T.; Fenger, C.; et al. NEXMIF encephalopathy: An X-linked disorder with male and female phenotypic patterns. Genet. Med. 2021, 23, 363–373. [Google Scholar] [CrossRef]
- Fan, H.C.; Lee, H.F.; Chi, C.S. SCN8A encephalopathy: Case report and literature review. Neurol. Int. 2021, 13, 143–150. [Google Scholar] [CrossRef]
- Crawford, K.; Xian, J.; Helbig, K.L.; Galer, P.D.; Parthasarathy, S.; Lewis-Smith, D.; Kaufman, M.C.; Fitch, E.; Ganesan, S.; O’Brien, M.; et al. Computational analysis of 10,860 phenotypic annotations in individuals with SCN2A-related disorders. Genet. Med. 2021, 23, 1263–1272. [Google Scholar] [CrossRef]
- Haddaway, N.R.; Collins, A.M.; Coughlin, D.; Kirk, S. The role of Google Scholar in evidence reviews and its applicability to grey literature searching. PLoS ONE 2015, 10, e0138237. [Google Scholar] [CrossRef] [PubMed]
- Hicks, D.; Wouters, P.; Waltman, L.; De Rijcke, S.; Rafols, I. Bibliometrics: The Leiden Manifesto for research metrics. Nat. News 2015, 520, 429. [Google Scholar] [CrossRef] [PubMed]
- Hens, K.; Peeters, H.; Dierickx, K. Genetic testing and counseling in the case of an autism diagnosis: A caregivers perspective. Eur. J. Med. Genet. 2016, 59, 452–458. [Google Scholar] [CrossRef] [PubMed]

| Cluster ID | Size | Silhouette | Mean Publication Year | LLR Label | Suggested Label |
|---|---|---|---|---|---|
| 0 | 218 | 0.823 | 2018 | Intellectual Disability | Networks and Pathways |
| 1 | 158 | 0.920 | 2019 | Gut Microbiota | Gut Microbiota |
| 2 | 132 | 0.850 | 2018 | Mouse Model | Fragile X Syndrome |
| 3 | 120 | 0.885 | 2018 | Mutant Mice | SHANK1,2,3 Genes |
| 4 | 119 | 0.831 | 2018 | Valproic Acid | Valproic Acid |
| 5 | 110 | 0.919 | 2019 | Genomic Architecture | Genomic Architecture |
| 6 | 106 | 0.825 | 2019 | Brain Organoid | Brain Organoid |
| 7 | 102 | 0.893 | 2020 | Psychiatric Disorder | Psychiatric Disorder |
| 8 | 72 | 0.905 | 2019 | Sex Difference | Sex Difference |
| 9 | 59 | 0.911 | 2018 | Autism Spectrum Disorder | Copy Number Variations (CNVs) |
| 10 | 49 | 0.985 | 2019 | Autistic Adult | Developmental Perspectives |
| 14 | 4 | 1.000 | 2020 | Antiseizure Drug | Antiseizure Drug |
| Reference | Citation Burstness | Publication Year | Burst Begin | Burst End | Duration | Betweenness Centrality | Sigma |
|---|---|---|---|---|---|---|---|
| Lord et al. [41] | 14.357 | 2018 | 2020 | 2022 | 2 | 0.0010 | 1.01 |
| Grove et al. [42] | 9.462 | 2019 | 2020 | 2022 | 2 | 0.0128 | 1.13 |
| Iakoucheva et al. [43] | 8.080 | 2019 | 2020 | 2022 | 2 | 0.0001 | 1.00 |
| Sharon et al. [45] | 7.827 | 2019 | 2020 | 2022 | 2 | 0.0066 | 1.05 |
| Ruzzo et al. [46] | 7.389 | 2019 | 2020 | 2022 | 2 | 0.0100 | 1.08 |
| Kim et al. [47] | 7.172 | 2011 | 2018 | 2019 | 1 | 0.0000 | 1.00 |
| Abraham et al. [48] | 6.816 | 2017 | 2020 | 2022 | 2 | 0.0031 | 1.02 |
| Lim et al. [49] | 6.702 | 2017 | 2019 | 2020 | 1 | 0.0003 | 1.00 |
| Yang et al. [50] | 6.693 | 2012 | 2018 | 2019 | 1 | 0.0013 | 1.01 |
| Lee et al. [51] | 6.311 | 2019 | 2020 | 2022 | 2 | 0.0006 | 1.00 |
| Nowakowski et al. [52] | 6.311 | 2017 | 2020 | 2022 | 2 | 0.0068 | 1.04 |
| Velmeshev et al. [53] | 6.311 | 2019 | 2020 | 2022 | 2 | 0.0013 | 1.01 |
| Matta et al. [54] | 6.311 | 2019 | 2020 | 2022 | 2 | 0.0011 | 1.01 |
| Estes and McAllister [55] | 6.214 | 2015 | 2018 | 2019 | 1 | 0.0020 | 1.01 |
| Goines and Ashwood [56] | 6.058 | 2013 | 2020 | 2022 | 2 | 0.0011 | 1.01 |
| Pantelis et al. [57] | 6.058 | 2014 | 2020 | 2022 | 2 | 0.0004 | 1.00 |
| Yuen et al. [58] | 5.975 | 2015 | 2018 | 2019 | 1 | 0.0008 | 1.01 |
| Antoine et al. [59] | 5.909 | 2019 | 2020 | 2022 | 2 | 0.0074 | 1.04 |
| Schafer et al. [60] | 5.805 | 2019 | 2020 | 2022 | 2 | 0.0031 | 1.02 |
| Stahl et al. [61] | 5.805 | 2019 | 2020 | 2022 | 2 | 0.0019 | 1.01 |
| Title | Coverage | Global Citing Score |
|---|---|---|
| Joensuu et al. [62] | 57 | 37 |
| Gandhi and Lee [67] | 52 | 9 |
| Guang et al. [71] | 47 | 98 |
| Garcia-Forn et al. [72] | 45 | 8 |
| Hui et al. [65] | 44 | 8 |
| Diaz-Caneja et al. [73] | 44 | 11 |
| Alonso-Gonzalez et al. [63] | 40 | 25 |
| DiCarlo and Wallace [74] | 40 | 2 |
| Eyring and Geschwind [66] | 39 | 5 |
| Iakoucheva et al. [43] | 39 | 94 |
| Title | Coverage | Global Citing Score |
|---|---|---|
| Guang et al. [71] | 38 | 98 |
| Patel et al. [85] | 37 | 9 |
| Yang and Shcheglovitov [86] | 33 | 10 |
| Panisi et al. [87] | 30 | 21 |
| Matta et al. [54] | 27 | 78 |
| Zheng et al. [75] | 26 | 8 |
| DiCarlo and Wallace [74] | 25 | 2 |
| Liu et al. [88] | 25 | 11 |
| Lombardo et al. [89] | 25 | 73 |
| Fattorusso et al. [77] | 25 | 140 |
| Title | Coverage | Global Citing Score |
|---|---|---|
| Wang et al. [94] | 64 | 8 |
| Verma et al. [91] | 46 | 23 |
| Gandhi and Lee [67] | 43 | 9 |
| Joensuu et al. [62] | 40 | 37 |
| Guang et al. [71] | 34 | 98 |
| Sungur et al. [93] | 34 | 12 |
| Bagni and Zukin [97] | 33 | 109 |
| Chaudry and Vasudevan [106] | 31 | 0 |
| Patel et al. [85] | 31 | 9 |
| Möhrle et al. [90] | 28 | 21 |
| Title | Coverage | Global Citing Score |
|---|---|---|
| Wang et al. [94] | 54 | 8 |
| Soler et al. [95] | 40 | 26 |
| Mossa et al. [103] | 35 | 19 |
| Yoo et al. [104] | 35 | 22 |
| Ali Rodriguez et al. [107] | 34 | 10 |
| Joensuu et al. [62] | 33 | 37 |
| Sungur et al. [93] | 31 | 12 |
| Yoo et al. [102] | 29 | 15 |
| Yang and Shcheglovitov [86] | 29 | 10 |
| Verma et al. [91] | 29 | 23 |
| Title | Coverage | Global Citing Score |
|---|---|---|
| St. Clair and Johnstone [108] | 22 | 13 |
| Tartaglione et al. [111] | 19 | 28 |
| Hui et al. [65] | 18 | 8 |
| Filice et al. [124] | 17 | 18 |
| Rylaarsdam and Guemez-Gamboa [14] | 16 | 118 |
| Napolitano et al. [125] | 16 | 0 |
| DiCarlo and Wallace [74] | 14 | 2 |
| Fink and Levine [112] | 14 | 14 |
| Patel et al. [85] | 14 | 9 |
| Nakai et al. [92] | 14 | 24 |
| Title | Coverage | Global Citing Score |
|---|---|---|
| Lord et al. [109] | 23 | 211 |
| Courchesne et al. [126] | 15 | 40 |
| Hoffmann et al. [127] | 15 | 14 |
| Ilieva et al. [115] | 14 | 39 |
| Hong et al. [128] | 12 | 31 |
| Chan et al. [116] | 12 | 12 |
| Niu and Parent [129] | 12 | 18 |
| Fetit et al. [130] | 11 | 6 |
| Griesi-Oliveira et al. [131] | 10 | 22 |
| Hui et al. [65] | 10 | 8 |
| Title | Coverage | Global Citing Score |
|---|---|---|
| Al-Dewik et al. [132] | 20 | 5 |
| Culotta and Penzes [141] | 18 | 12 |
| Breen et al. [133] | 14 | 13 |
| Gordon and Geschwind [142] | 13 | 7 |
| Prem et al. [143] | 13 | 7 |
| Muhle et al. [144] | 12 | 76 |
| Grabrucker [145] | 14 | 2 |
| Saxena et al. [146] | 12 | 5 |
| Scuderi and Verkhratsky [147] | 11 | 8 |
| Fink and Levine [112] | 11 | 14 |
| Title | Coverage | Global Citing Score |
|---|---|---|
| Lord et al. [41] | 18 | 211 |
| Park et al. [157] | 16 | 23 |
| Jiang et al. [158] | 15 | 0 |
| Urresti et al. [159] | 12 | 11 |
| Walker et al. [160] | 12 | 62 |
| Willsey et al. [161] | 12 | 1 |
| Hoffmann et al. [127] | 12 | 14 |
| Sullivan and Geschwind [162] | 12 | 156 |
| Rees and Owen [163] | 12 | 28 |
| Mullins et al. [164] | 11 | 94 |
| Title | Coverage | Global Citing Score |
|---|---|---|
| Rylaarsdam and Guemez-Gamboa [14] | 11 | 118 |
| Lord et al. [41] | 10 | 211 |
| Napolitano et al. [125] | 9 | 0 |
| Rujeedawa and Zaman [165] | 9 | 0 |
| Lai et al. [171] | 8 | 53 |
| Kallitsounaki and Williams [169] | 6 | 0 |
| Müller and Fishman [172] | 6 | 47 |
| Wilson et al. [173] | 6 | 7 |
| Howes et al. [174] | 6 | 105 |
| Yuen et al. [175] | 6 | 12 |
| Title | Coverage | Global Citing Score |
|---|---|---|
| Jønch et al. [176] | 11 | 19 |
| Egolf et al. [179] | 11 | 14 |
| Deshpande and Weiss [190] | 11 | 22 |
| Lengyel et al. [180] | 10 | 1 |
| Takumi and Tamada [191] | 9 | 55 |
| Rylaarsdam and Guemez-Gamboa [14] | 9 | 118 |
| Kushima et al. [192] | 8 | 114 |
| Bristow et al. [181] | 7 | 11 |
| Pucilowska et al. [182] | 7 | 39 |
| Campbell and Granato [149] | 7 | 3 |
| Title | Coverage | Global Citing Score |
|---|---|---|
| Zwaigenbaum et al. [193] | 7 | 20 |
| Barros et al. [203] | 6 | 2 |
| Al-Dewik et al. [132] | 6 | 5 |
| Lacroix et al. [204] | 6 | 1 |
| Kirst et al. [196] | 5 | 0 |
| Hollin [205] | 5 | 0 |
| Nebel et al. [206] | 5 | 0 |
| Belcher et al. [199] | 5 | 0 |
| Rozenblatt-Perkal and Zaidman-Zait [197] | 5 | 1 |
| McCracken et al. [207] | 5 | 5 |
| Title | Coverage | Global Citing Score |
|---|---|---|
| Hakami [208] | 4 | 2 |
| Hakami [209] | 4 | 3 |
| Stamberger et al. [219] | 3 | 10 |
| Hawkins et al. [211] | 3 | 5 |
| Fan et al. [220] | 2 | 4 |
| De Maria et al. [212] | 2 | 4 |
| Crawford et al. [221] | 2 | 5 |
| Galanopoulou et al. [214] | 2 | 6 |
| Cali et al. [213] | 2 | 0 |
| Raga et al. [215] | 2 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, M.; Carollo, A.; Dimitriou, D.; Esposito, G. Recent Developments in Autism Genetic Research: A Scientometric Review from 2018 to 2022. Genes 2022, 13, 1646. https://doi.org/10.3390/genes13091646
Lim M, Carollo A, Dimitriou D, Esposito G. Recent Developments in Autism Genetic Research: A Scientometric Review from 2018 to 2022. Genes. 2022; 13(9):1646. https://doi.org/10.3390/genes13091646
Chicago/Turabian StyleLim, Mengyu, Alessandro Carollo, Dagmara Dimitriou, and Gianluca Esposito. 2022. "Recent Developments in Autism Genetic Research: A Scientometric Review from 2018 to 2022" Genes 13, no. 9: 1646. https://doi.org/10.3390/genes13091646
APA StyleLim, M., Carollo, A., Dimitriou, D., & Esposito, G. (2022). Recent Developments in Autism Genetic Research: A Scientometric Review from 2018 to 2022. Genes, 13(9), 1646. https://doi.org/10.3390/genes13091646





