Study on the Characteristics of Coarse Feeding Tolerance of Ding’an Pigs: Phenotypic and Candidate Genes Identification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animal Feeding Management and Sample Collection
2.2. Collection of Phenotypes
2.2.1. Measurement of Growth Performance
2.2.2. Measurement of Apparent Digestibility of Crude Fiber
2.2.3. Measurement of Carcass Traits
2.2.4. Measurement of Meat Quality Traits
2.2.5. Measurement of Serum Biochemical Indicators
2.3. Total RNA Extraction and Transcriptome Sequencing
2.4. Principal Component Analysis
2.5. Differential Gene Analysis and Validation of Transcriptome Data Accuracy
2.6. Functional Analysis of DEGs
2.7. Weighted Gene Co-Expression Network Analysis
2.8. Network of Differential Gene-Protein Interactions
2.9. Statistical Analysis of Data
3. Results
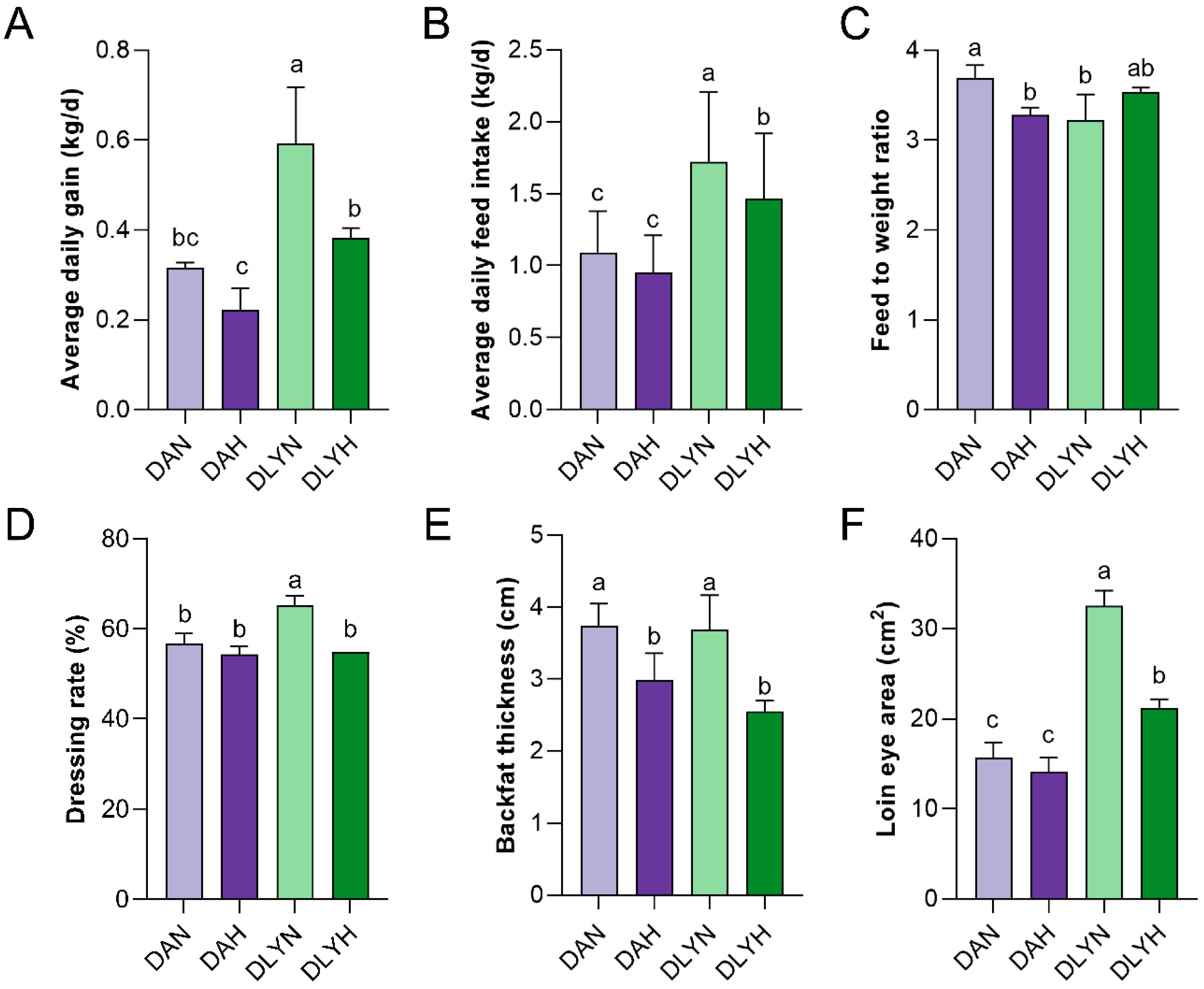
3.1. Differential Phenotypic Collection between DA (Ding’an) Pigs and DLY (Duroc-Landrace-Yorkshire) Pigs
3.2. Inter-Phenotypic Correlation Analysis
3.3. Analysis and Validation of Differentially Expressed Genes (DEGs)
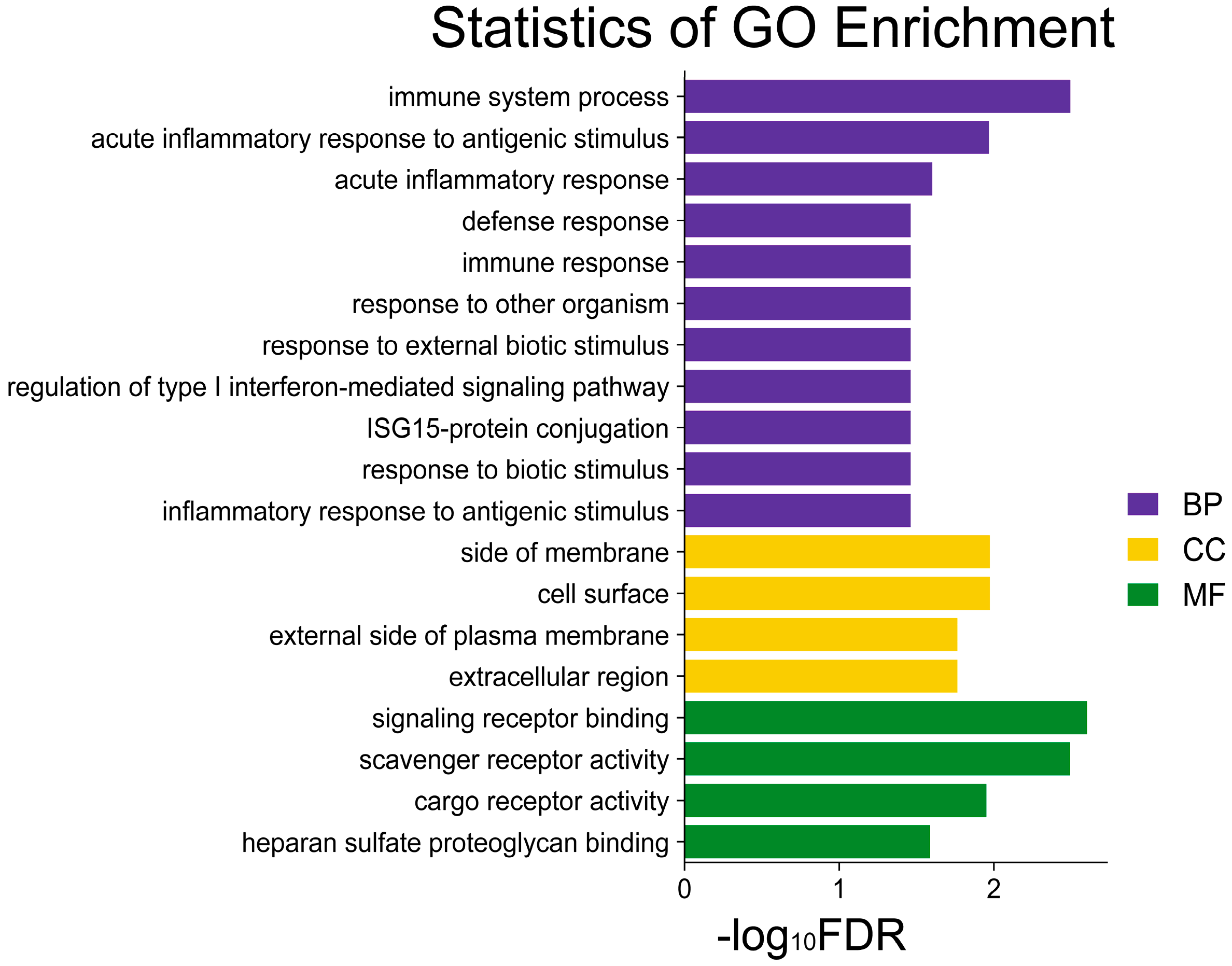
3.4. Functional Enrichment Analysis of Candidate DEGs for Coarse Feeding Tolerance
3.5. Weighted Gene Co-Expression Network Analysis (WGCNA) of All Expressed Genes
3.6. Identification of Hub DEGs for Coarse Feeding Tolerance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Liu, H.; Wang, W.; Huang, L. Polymorphic analysis of TLR2 gene in native Hainan pig breeds based on sequencing technology. Anim. Husb. Feed. Sci. 2019, 11, 135–139. [Google Scholar]
- Diao, S.Q.; Xu, Z.T.; Ye, S.P.; Huang, S.W.; Teng, J.J.; Yuan, X.L.; Chen, Z.M.; Zhang, H.; Li, J.Q.; Zhang, Z. Exploring the genetic features and signatures of selection in South China indigenous pigs. J. Integr. Agric. 2021, 20, 1359–1371. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Y.; Wang, W.; Huang, L. Polymorphic analysis of TLR4 gene in Hainan local pig breeds based on sequencing technology. Anim. Husb. Feed Sci. 2020, 12, 1–5. [Google Scholar]
- Wang, F.F.; Zha, Z.L.; He, Y.Z.; Li, J.; Zhong, Z.Q.; Xiao, Q.; Tan, Z. Genome-Wide Re-Sequencing Data Reveals the Population Structure and Selection Signatures of Tunchang Pigs in China. Animals 2023, 13, 1835. [Google Scholar] [CrossRef] [PubMed]
- Ao, X.; Zhou, J.; Li, P.; Diao, H.; Li, S.; Tang, W.; He, X.; Yan, H.; Liu, J. Comparative study on digestibility of fiber feed ingredients between Chuanzang black pigs and Duroc*Landrace*Yorkshire pigs. Chin. J. Anim. Nutr. 2023, 35, 3555–3562. [Google Scholar]
- Kim, M.; Roura, E.; Choi, Y.; Kim, J. Transcriptomic Analysis of the Porcine Gut in Response to Heat Stress and Dietary Soluble Fiber from Beet Pulp. Genes 2022, 13, 1456. [Google Scholar] [CrossRef] [PubMed]
- Pu, G.; Hou, L.M.; Du, T.R.; Wang, B.B.; Liu, H.; Li, K.J.; Niu, P.P.; Zhou, W.D.; Huang, R.H.; Li, P.H. Effects of short-term feeding with high fiber diets on growth, utilization of dietary fiber, and microbiota in pigs. Front. Microbiol. 2022, 13, 963917. [Google Scholar] [CrossRef] [PubMed]
- Du, T.R.; Li, P.H.; Niu, Q.; Pu, G.; Wang, B.B.; Liu, G.S.; Li, P.H.; Niu, P.P.; Zhang, Z.P.; Wu, C.W.; et al. Effects of Varying Levels of Wheat Bran Dietary Fiber on Growth Performance, Fiber Digestibility and Gut Microbiota in Erhualian and Large White Pigs. Microorganisms 2023, 11, 2474. [Google Scholar] [CrossRef] [PubMed]
- Noblet, J.; Wu, S.B.; Choct, M. Methodologies for energy evaluation of pig and poultry feeds: A review. Anim. Nutr. 2022, 8, 185–203. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, C.; Niu, J.; Cui, Z.X.; Zhao, X.Y.; Li, W.X.; Zhang, Y.W.; Yang, Y.; Gao, P.F.; Guo, X.H.; et al. Impacts of dietary fiber level on growth performance, apparent digestibility, intestinal development, and colonic microbiota and metabolome of pigs. J. Anim. Sci. 2023, 101, skad174. [Google Scholar] [CrossRef]
- Jin, S.S.; Wijerathne, C.U.B.; Au-Yeung, K.K.W.; Lei, H.G.; Yang, C.B.; Karmin, O. Effects of high- and low-fiber diets on intestinal oxidative stress in growing-finishing pigs. J. Anim. Sci. 2022, 100, skac306. [Google Scholar] [CrossRef] [PubMed]
- Garrido, N.; Albuquerque, A.; Charneca, R.; Costa, F.; Marmelo, C.; Ramos, A.; Martin, L.; Martins, J.M. Transcriptomic Profiling of Subcutaneous Backfat in Castrated and Intact Alentejano Pigs Finished Outdoors with Commercial and Fiber-Rich Diets. Genes 2023, 14, 1722. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, X.Y.; Zhao, G.S.; Xue, H.P.; Wang, Y.P.; Ren, Y.F.; Li, J.X.; Wang, H.Z.; Wang, J.Y.; Song, Q.Y. Comparative Analysis of Structural Composition and Function of Intestinal Microbiota between Chinese Indigenous Laiwu Pigs and Commercial DLY Pigs. Vet. Sci. 2023, 10, 524. [Google Scholar] [CrossRef]
- Yang, L.J.; Wang, G.; Zhou, J.Y.; Yang, Y.T.; Pan, H.B.; Zeng, X.F.; Qiao, S.Y. Exploration of the Potential for Efficient Fiber Degradation by Intestinal Microorganisms in Diqing Tibetan Pigs. Fermentation 2021, 7, 275. [Google Scholar] [CrossRef]
- Li, X.J.; Lu, L.Y.; Tong, X.W.; Li, R.D.; Jin, E.; Ren, M.; Gao, Y.F.; Gu, Y.F.; Li, S.H. Transcriptomic Profiling of Meat Quality Traits of Skeletal Muscles of the Chinese Indigenous Huai Pig and Duroc Pig. Genes 2023, 14, 1548. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, A.; Ovilo, C.; Núñez, Y.; Benítez, R.; López-Garcia, A.; García, F.; Félix, M.D.; Laranjo, M.; Charneca, R.; Martins, J.M. Comparative Transcriptomic Analysis of Subcutaneous Adipose Tissue from Local Pig Breeds. Genes 2020, 11, 422. [Google Scholar] [CrossRef]
- Lange, K.; Hugenholtz, F.; Jonathan, M.C.; Schols, H.A.; Kleerebezem, M.; Smidt, H.; Mueller, M.; Hooiveld, G.J.E.J. Comparison of the effects of five dietary fibers on mucosal transcriptional profiles, and luminal microbiota composition and SCFA concentrations in murine colon. Mol. Nutr. Food Res. 2015, 59, 1590–1602. [Google Scholar] [CrossRef]
- Tang, W.D.; Kang, J.C.; Sun, R.P.; Chao, Z.; Liu, H.L.; Huang, L.L.; Wang, F. Comparative study of meat quality characteristics of Hainan Ding’an pigs and their F1 generation crossed with Duroc. Swine Sci. 2023, 40, 122–124. (In Chinese) [Google Scholar]
- Xue, P.X.; Luo, Y.B.; Song, Y.X. Colonic Microbiota Improves Fiber Digestion Ability and Enhances Absorption of SCFAs in Hainan Local Pigs, BMC Microbiology 2023, PREPRINT (Version 1). Res. Sq. 2023, preprint. Available online: https://www.researchsquare.com/article/rs-2952520/v1 (accessed on 31 May 2023). [CrossRef]
- Kim, B.G.; Tian, J.Z.; Lim, J.S.; Kil, D.Y.; Jeon, H.Y.; Chung, Y.K.; Kim, Y.Y. Influences of enzyme complex supplementation on growth, ileal and apparent fecal digestibility and morphology of small intestine in pigs. Asian Australas. J. Anim. Sci. 2004, 17, 1729–1735. [Google Scholar] [CrossRef]
- GB/T 6434-2022; Determination of Crude Fiber Content in Feed. National Feed Industry Standardisation Technical Committee: Beijing, China, 2022.
- NY/T 825-2004; Technical Specifications for Determining the Carcass Traits of Lean Meat Pigs. Industry Standards—Agriculture: Guangzhou, China, 2004.
- Wu, L.T.; Tan, L.M.; You, C.Y.; Lan, T.Y.; Li, W.X.; Xu, Y.T.; Ren, Z.X.; Ding, Q.; Zhou, C.Y.; Tang, Z.R.; et al. Effects of dietary niacinamide and CP concentrations on the nitrogen excretion, growth performance, and meat quality of pigs. Animal 2023, 17, 100869. [Google Scholar] [CrossRef] [PubMed]
- NY/T 821-2019; Technical Regulations for Determining the Quality of Pork. Ministry of Agriculture and Rural Development: Beijing, China, 2019.
- Wang, Y.; Zhang, H.; Yan, E.; He, L.; Guo, J.; Zhang, X.; Yin, J. Carcass and meat quality traits and their relationships in Duroc × Landrace × Yorkshire barrows slaughtered at various seasons. Meat Sci. 2023, 198, 109117. [Google Scholar] [CrossRef] [PubMed]
- GB 5009.6-2016; National Standard for Food Safety-Determination of Fat in Food. National Health and Family Planning Commission of the People’s Republic of China; State Food and Drug Administration: Beijing, China, 2016.
- Wang, S.J.; Wang, B.X.; He, H.; Sung, A.M.; Guo, C.H. A new set of reference housekeeping genes for the normalization RT-qPCR data from the intestine of piglets during weaning. PLoS ONE 2018, 13, e0204583. [Google Scholar] [CrossRef] [PubMed]
- Barreto Sanchez, A.L.; Wang, Q.; Thiam, M.; Wang, Z.; Zhang, J.; Zhang, Q.; Zhang, N.; Li, Q.; Wen, J.; Zhao, G. Liver Transcriptome Response to Heat Stress in Beijing You Chickens and Guang Ming Broilers. Genes 2022, 13, 416. [Google Scholar] [CrossRef] [PubMed]
- Diao, S.; Huang, S.; Chen, Z.; Teng, J.; Ma, Y.; Yuan, X.; Chen, Z.; Zhang, H.; Li, J.; Zhang, Z. Genome-Wide Signatures of Selection Detection in Three South China Indigenous Pigs. Genes 2019, 10, 346. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.Q.; Wang, Z.Y.; Xie, X.F.; Tian, S.S.; Wang, F.F.; Wang, Q.S.; Ni, S.H.; Pan, Y.C.; Xiao, Q. Evaluation of the Genetic Diversity, Population Structure and Selection Signatures of Three Native Chinese Pig Populations. Animals 2023, 13, 2010. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Cheng, Y.; Azad, M.A.K.; Dong, H.; He, J.; Huang, P.; Kong, X. Dietary fiber alters immunity and intestinal barrier function of different breeds of growing pigs. Front. Immunol. 2023, 14, 1104837. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yin, J.; Tan, B.; Chen, J.S.; Zhang, H.H.; Li, Z.Q.; Ma, X.K. Physiological function and application of dietary fiber in pig nutrition: A review. Anim. Nutr. 2021, 7, 259–267. [Google Scholar] [CrossRef]
- Bekebrede, A.F.; Noorman, L.; Keijer, J.; de Boer, V.C.J.; Gerrits, W.J.J. Functional metabolic capacity of pig colonocytes is differentially modulated by fermentable fibre and poorly digestible protein. Animal 2022, 16, 100625. [Google Scholar] [CrossRef]
- Chen, T.T.; Chen, D.W.; Tian, G.; Zheng, P.; Mao, X.B.; Yu, J.; He, J.; Huang, Z.Q.; Luo, Y.H.; Luo, J.Q.; et al. Effects of soluble and insoluble dietary fiber supplementation on growth performance, nutrient digestibility, intestinal microbe and barrier function in weaning piglet. Anim. Feed Sci. Technol. 2020, 260, 114335. [Google Scholar] [CrossRef]
- Luo, Y.H.; He, J.; Li, H.; Lan, C.; Cai, J.Y.; Chen, H.; Tian, G.; Wang, H.F.; Wang, Q.Y.; He, J.; et al. Wheat bran fermented by mixed fungal strains improves the digestibility of crude fiber and may benefit the gut health without impacting the growth performance in weaned pigs. Food Funct. 2021, 12, 2962–2971. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Mao, X.B.; Che, L.Q.; Yu, B.; He, J.; Yu, J.; Han, G.Q.; Huang, Z.Q.; Zheng, P.; Chen, D.W. Impact of fiber types on gut microbiota, gut environment and gut function in fattening pigs. Anim. Feed Sci. Technol. 2014, 195, 101–111. [Google Scholar] [CrossRef]
- Chen, T.; Chen, D.; Tian, G.; Zheng, P.; Mao, X.; Yu, J.; He, J.; Huang, Z.; Luo, Y.; Luo, J.; et al. Soluble Fiber and Insoluble Fiber Regulate Colonic Microbiota and Barrier Function in a Piglet Model. Biomed Res. Int. 2019, 7809171. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Shi, K.; Wang, Y.; Zou, D.; Guo, S.; Li, T.; Xu, H.; Ma, X.; Liu, J.; Song, H.; et al. Effect of Huangqin Tang on Colonic Gene Expression in Rats with Ulcerative Colitis. Int. J. Genom. 2020, 2020, 4238757. [Google Scholar] [CrossRef] [PubMed]
- Yde, J.; Wu, Q.; Borg, J.F.; Fenton, R.A.; Moeller, H.B. A systems-level analysis of bile acids effects on rat colon epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2022, 322, G34–G48. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.D.; Ramkumar, S.; Golczak, M.; von Lintig, J. Genetic deletion of Bco2 and Isx establishes a golden mouse model for carotenoid research. Mol. Metab. 2023, 73, 101742. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Hong, J.H. Trafficking of carbonic anhydrase 12 and bicarbonate transporters by histamine stimulation mediates intracellular acidic scenario in lung cancer cells. J. Enzym. Inhib. Med. Chem. 2023, 38, 2247181. [Google Scholar] [CrossRef]
- Michl, J.; White, B.; Monterisi, S.; Bodmer, W.F.; Swietach, P. Phenotypic screen of sixty-eight colorectal cancer cell lines identifies CEACAM6 and CEACAM5 as markers of acid resistance. Proc. Natl. Acad. Sci. USA 2024, 121, e2319055121. [Google Scholar] [CrossRef]
- Zimmer, B.M.; Barycki, J.J.; Simpson, M.A. Integration of Sugar Metabolism and Proteoglycan Synthesis by UDP-glucose Dehydrogenase. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2021, 69, 13–23. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, N.; Xing, J.; Zhang, S.; Chen, Y.; Xu, D.; Gu, J. UDP-glucuronate metabolism controls RIPK1-driven liver damage in nonalcoholic steatohepatitis. Nat. Commun. 2023, 14, 2715. [Google Scholar] [CrossRef]
- Harrington, B.S.; Kamdar, R.; Ning, F.; Korrapati, S.; Caminear, M.W.; Hernandez, L.F.; Butcher, D.; Edmondson, E.F.; Traficante, N.; Hendley, J.; et al. UGDH promotes tumor-initiating cells and a fibroinflammatory tumor microenvironment in ovarian cancer. J. Exp. Clin. Cancer Res. CR 2023, 42, 270. [Google Scholar] [CrossRef]
- Chandrasekaran, R.; Mathieu, C.; Sheth, R.; Cheng, A.P.; Fong, D.; McCormack, R.; El-Gabalawy, H.; Alishetty, S.; Paige, M.; Hoemann, C.D. UDP-glucose dehydrogenase (UGDH) activity is suppressed by peroxide and promoted by PDGF in fibroblast-like synoviocytes: Evidence of a redox control mechanism. PLoS ONE 2022, 17, e0274420. [Google Scholar] [CrossRef] [PubMed]
- Zhan, D.; Yalcin, F.; Ma, D.; Fu, Y.; Wei, S.; Lal, B.; Li, Y.; Dzaye, O.; Laterra, J.; Ying, M.; et al. Targeting UDP-α-d-glucose 6-dehydrogenase alters the CNS tumor immune microenvironment and inhibits glioblastoma growth. Genes Dis. 2022, 9, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Yan, Y.; Shen, C. A Prognostic Pyroptosis-Related lncRNAs Risk Model Correlates With the Immune Microenvironment in Colon Adenocarcinoma. Front. Cell Dev. Biol. 2021, 9, 811734. [Google Scholar] [CrossRef] [PubMed]
- Rui, S.; Wang, D.; Huang, Y.; Xu, J.; Zhou, H.; Zhang, H. Prognostic value of SLC4A4 and its correlation with the microsatellite instability in colorectal cancer. Front. Oncol. 2023, 13, 1179120. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zeng, J.; Lin, D.; Xia, H.; Wang, X.; Chen, L.; Chen, H.; Huang, L.; Zeng, C. Extracellular vesicles derived from cancer-associated fibroblast carries miR-224-5p targeting SLC4A4 to promote the proliferation, invasion and migration of colorectal cancer cells. Carcinogenesis 2021, 42, 1143–1153. [Google Scholar] [CrossRef] [PubMed]
- Henriques, S.N.; Oliveira, L.; Santos, R.F.; Carmo, A.M. CD6-mediated inhibition of T cell activation via modulation of Ras. Cell Commun. Signal. CCS 2022, 20, 184. [Google Scholar] [CrossRef] [PubMed]
- Casadó-Llombart, S.; Velasco-de Andrés, M.; Català, C.; Leyton-Pereira, A.; Gutiérrez-Cózar, R.; Suárez, B.; Armiger, N.; Carreras, E.; Esteller, M.; Ricart, E.; et al. Experimental and genetic evidence for the impact of CD5 and CD6 expression and variation in inflammatory bowel disease. Front. Immunol. 2022, 13, 966184. [Google Scholar] [CrossRef] [PubMed]
- Ling, A.; Löfgren-Burström, A.; Larsson, P.; Li, X.R.; Wikberg, M.L.; Öberg, Å.; Stenling, R.; Edin, S.; Palmqvist, R. TAP1 down-regulation elicits immune escape and poor prognosis in colorectal cancer. Oncoimmunology 2017, 6, e1356143. [Google Scholar] [CrossRef]
- Theodoratos, A.; Whittle, B.; Enders, A.; Tscharke, D.C.; Roots, C.M.; Goodnow, C.C.; Fahrer, A.M. Mouse strains with point mutations in TAP1 and TAP2. Immunol. Cell Biol. 2010, 88, 72–78. [Google Scholar] [CrossRef]
- Johari, M.; Vihola, A.; Palmio, J.; Jokela, M.; Jonson, P.H.; Sarparanta, J.; Huovinen, S.; Savarese, M.; Hackman, P.; Udd, B. Comprehensive transcriptomic analysis shows disturbed calcium homeostasis and deregulation of T lymphocyte apoptosis in inclusion body myositis. J. Neurol. 2022, 269, 4161–4173. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Gomes, D.; Yuan, F.; Feng, J.; Zhang, X.; O’Toole, T.E. Oral Polystyrene Consumption Potentiates Atherosclerotic Lesion Formation in ApoE(−/−) Mice. Circ. Res. 2024, 134, 1228–1230. [Google Scholar] [CrossRef]
- Hutchison, E.R.; Kasahara, K.; Zhang, Q.; Vivas, E.I.; Cross, T.L.; Rey, F.E. Dissecting the impact of dietary fiber type on atherosclerosis in mice colonized with different gut microbial communities. NPJ Biofilms Microbiomes 2023, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.; Dou, P.; Cai, W.; Mao, M.; Wu, R. MiR-181a-5p promotes osteogenesis by targeting BMP3. Aging 2023, 15, 734–747. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, L.; Kang, S.; Gnanaprakasam, J.R.; Wang, R. The lactate dehydrogenase (LDH) isoenzyme spectrum enables optimally controlling T cell glycolysis and differentiation. Sci. Adv. 2023, 9, eadd9554. [Google Scholar] [CrossRef]
- Cheng, A.; Zhang, P.; Wang, B.; Yang, D.; Duan, X.; Jiang, Y.; Xu, T.; Jiang, Y.; Shi, J.; Ding, C.; et al. Aurora-A mediated phosphorylation of LDHB promotes glycolysis and tumor progression by relieving the substrate-inhibition effect. Nat. Commun. 2019, 10, 5566. [Google Scholar] [CrossRef] [PubMed]
- Lattier, J.M.; De, A.; Chen, Z.; Morales, J.E.; Lang, F.F.; Huse, J.T.; McCarty, J.H. Megalencephalic leukoencephalopathy with subcortical cysts 1 (MLC1) promotes glioblastoma cell invasion in the brain microenvironment. Oncogene 2020, 39, 7253–7264. [Google Scholar] [CrossRef] [PubMed]
- De, A.; Lattier, J.M.; Morales, J.E.; Kelly, J.R.; Zheng, X.; Chen, Z.; Sebastian, S.; Nassiri Toosi, Z.; Huse, J.T.; Lang, F.F.; et al. Glial Cell Adhesion Molecule (GlialCAM) Determines Proliferative versus Invasive Cell States in Glioblastoma. J. Neurosci. Off. J. Soc. Neurosci. 2023, 43, 8043–8057. [Google Scholar] [CrossRef]
- Ma, C.; Wu, S.; Li, N.; Chen, Y.; Yan, K.; Li, Z.; Zheng, L.; Lei, J.; Woolford, J.L., Jr.; Gao, N. Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat. Struct. Mol. Biol. 2017, 24, 214–220. [Google Scholar] [CrossRef]
- Yang, L.; Dong, Z.; Li, S.; Chen, T. ESM1 promotes angiogenesis in colorectal cancer by activating PI3K/Akt/mTOR pathway, thus accelerating tumor progression. Aging 2023, 15, 2920–2936. [Google Scholar] [CrossRef]
- Lu, J.; Liu, Q.; Zhu, L.; Liu, Y.; Zhu, X.; Peng, S.; Chen, M.; Li, P. Endothelial cell-specific molecule 1 drives cervical cancer progression. Cell Death Dis. 2022, 13, 1043. [Google Scholar] [CrossRef] [PubMed]




| GS | p.GS | MM | p.MM | Hub Gene | Trait |
|---|---|---|---|---|---|
| 0.91 | 1.11 × 10−6 | −0.97 | 1.46 × 10−9 | LDHB | CF |
| 0.75 | 0.00079 | −0.97 | 1.46 × 10−9 | LDHB | Intramuscular_fat |
| 0.69 | 0.00274 | −0.97 | 1.46 × 10−9 | LDHB | Marbling_score |
| −0.90 | 1.81 × 10−6 | −0.97 | 1.46 × 10−9 | LDHB | Shear_force |
| 0.80 | 0.00017 | −0.97 | 1.46 × 10−9 | LDHB | GLU |
| −0.80 | 0.00019 | 0.78 | 0.00034 | MLC1 | CF |
| 0.71 | 0.00209 | 0.78 | 0.00034 | MLC1 | Shear_force |
| −0.65 | 0.00631 | 0.78 | 0.00038 | LSG1 | CF |
| 0.86 | 0.00002 | 0.78 | 0.00038 | LSG1 | Shear_force |
| −0.62 | 0.00996 | 0.78 | 0.00038 | LSG1 | GLU |
| −0.61 | 0.01152 | 0.69 | 0.00294 | ESM1 | CF |
| 0.73 | 0.00120 | 0.69 | 0.00294 | ESM1 | Shear_force |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.; Xue, M.; Wang, F.; Tang, Q.; Luo, Y.; Zheng, M.; Wang, Y.; Xue, P.; Dong, N.; Sun, R.; et al. Study on the Characteristics of Coarse Feeding Tolerance of Ding’an Pigs: Phenotypic and Candidate Genes Identification. Genes 2024, 15, 599. https://doi.org/10.3390/genes15050599
Song Y, Xue M, Wang F, Tang Q, Luo Y, Zheng M, Wang Y, Xue P, Dong N, Sun R, et al. Study on the Characteristics of Coarse Feeding Tolerance of Ding’an Pigs: Phenotypic and Candidate Genes Identification. Genes. 2024; 15(5):599. https://doi.org/10.3390/genes15050599
Chicago/Turabian StyleSong, Yanxia, Mingming Xue, Feng Wang, Qiguo Tang, Yabiao Luo, Meili Zheng, Yubei Wang, Pengxiang Xue, Ningqi Dong, Ruiping Sun, and et al. 2024. "Study on the Characteristics of Coarse Feeding Tolerance of Ding’an Pigs: Phenotypic and Candidate Genes Identification" Genes 15, no. 5: 599. https://doi.org/10.3390/genes15050599





